CS224W-Machine Learning with Graph-Deep Generative Models for Graphs
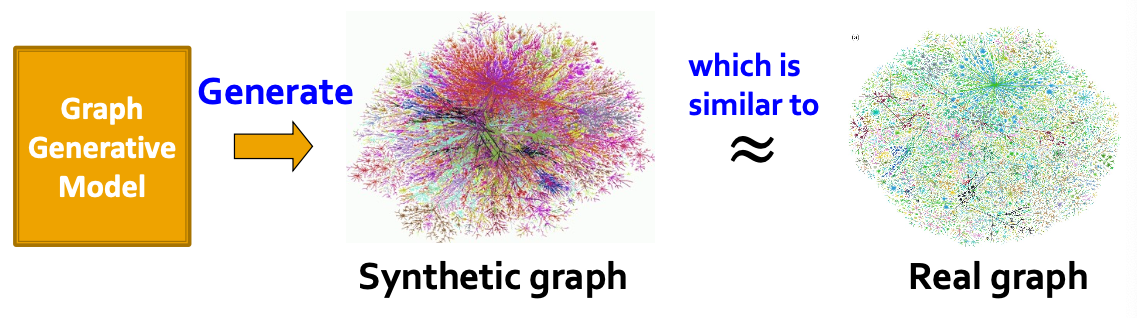
Graph Generation
- But how are these graphs generated?
- We want to generate realistic graphs, using graph generative modes
- Applications:
- Drug discovery, material design
- Social network modeling
- History of Graph Generation
- Step 1: Properties of real-world graphs
- A successful graph generative model should fit these peroperties.
- Step 2: Traditional graph generative models
- Each come with different assumptions on the graph formulation process
- Step 3: Deep graph generative models
- Learn the graph formation process from the data.
- Step 1: Properties of real-world graphs
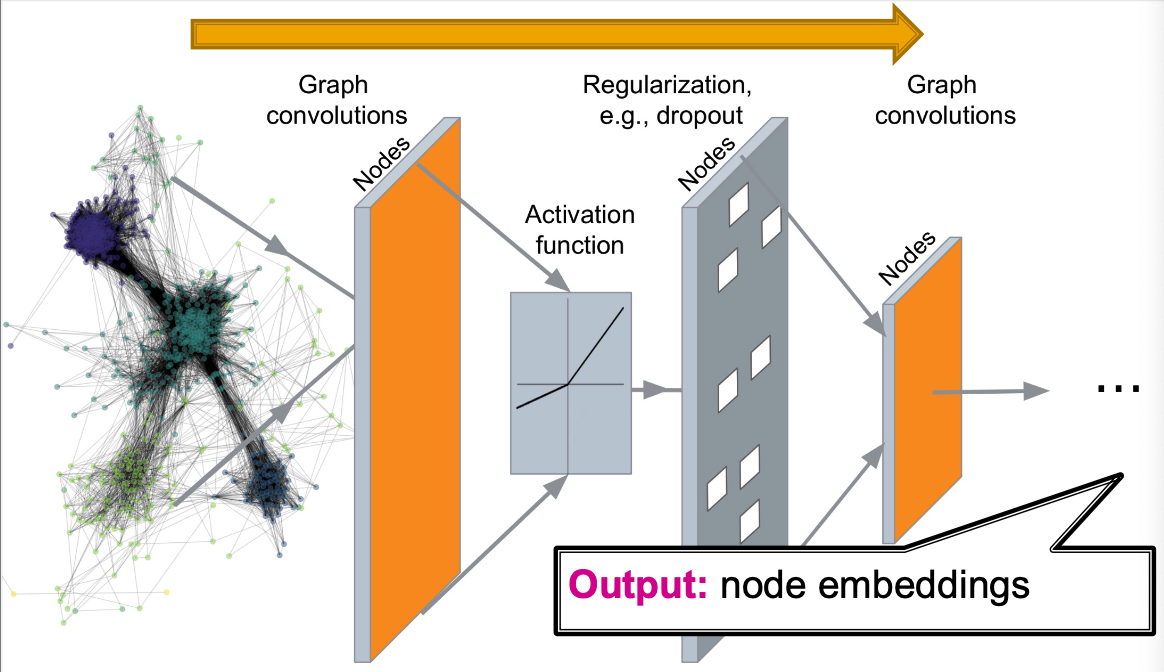
- Deep Graph Encoders
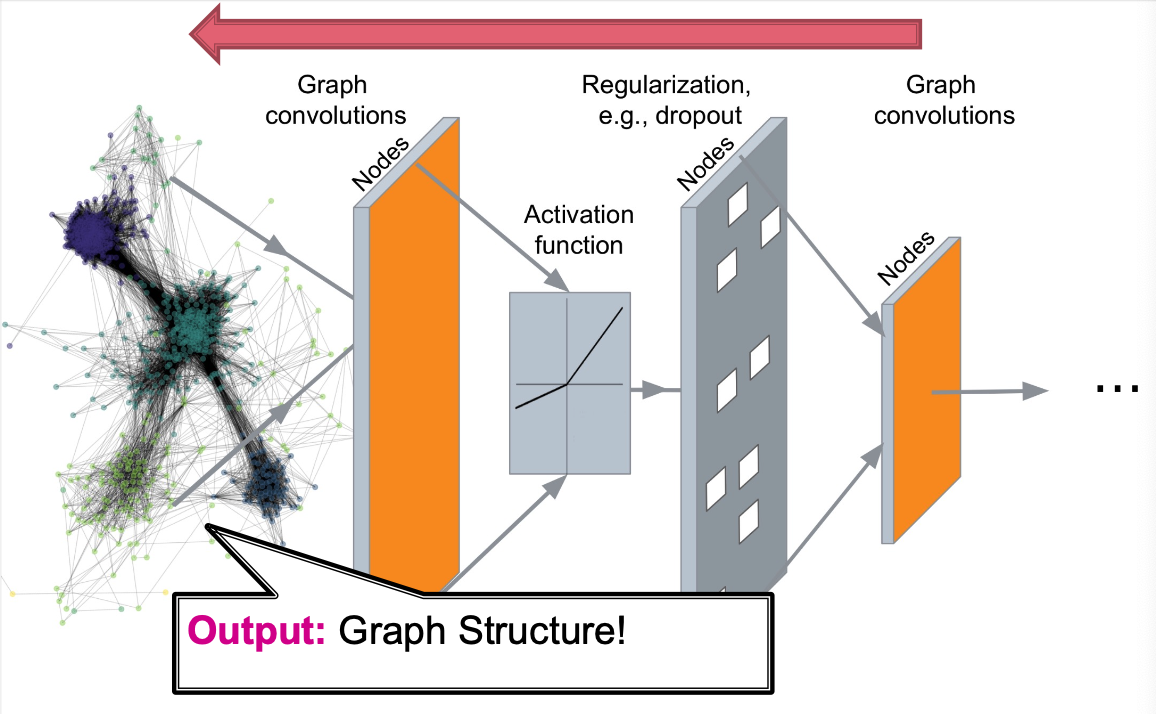
- Deep Graph Decoders (this lecture)
Machine Learning for Graph Generation
- Task 1: Realistic graph generation
- Generate graphs that are similar to a given set of graphs
- Task 2: Goal-directed graph generation
- Generate graphs that optimize given objectives / constraints
- E.g., Drug molecule generations / optimization
- Generate graphs that optimize given objectives / constraints
Graph Generative Models
- Given: Graphs sampled from .
- Goal:
- Learn the distribution .
- Sample from

Generative Models Basics
Setup:
- Assume we want to learn a generative model from a set of data points (i.e., graphs)
- is the data distribution, which is never known to us, but have sampled
- is the model, parameterized by , that we use to approximate .
- Goal:
- Make close to (Density estimation)
- Key Principle: maximum Likelihood
- Fundamental approach to modeling distributions
- Find parameters , such that for observed data points the has the highest value, among all possible choices of
- That is, find the model that is most likely to have generated the observed data .
- Find parameters , such that for observed data points the has the highest value, among all possible choices of
- Make sure we can sample from (Sampling)
- We need to generate examples (graphs) from
- Goal: Sample from a complex distribution
- The most common approach:
- Sample from a simple noise distribution
- Transform the noise via
- How too design ?
- Use Deep Neural Networks, and train it using the data we have!
- Make close to (Density estimation)
Deep Generative Models
Auto-regressive models:
- is used for both density estimation and sampling (remember out two goals)
- Other models like Variational Auto Encoders(VAEs), Generative Adversarial Nets (GAN) have 2 or more models, each playing one of the roles
- Idea: Chain rule. Joint distribution is a product of conditional distributions:
- E.g., is a vector, is the -th dimension; is a sentence, is the -th word.
- In our case: will be the -th action (add node, add edge)
GraphRNN: Generating Realistic Graphs
Generating graphs via sequentially adding nodes and edges
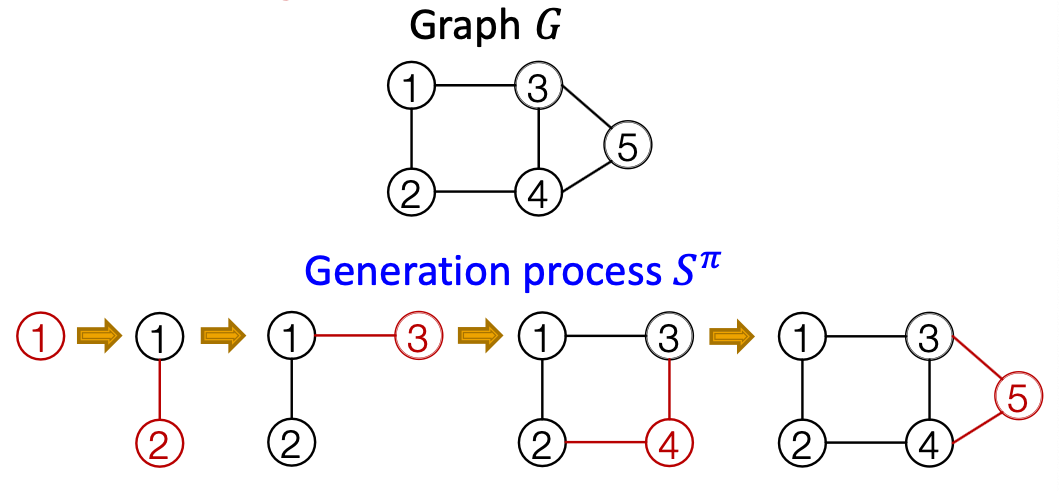
Model Graphs as Sequences
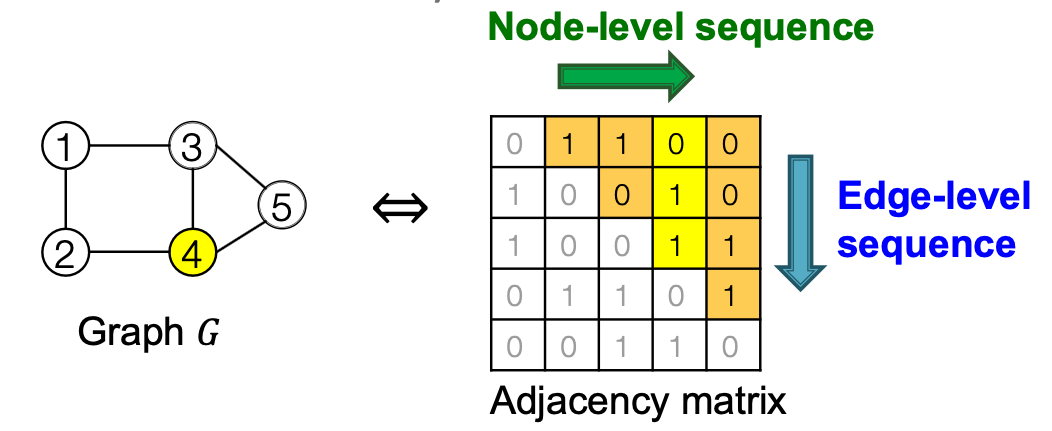
Graph with node ordering can be uniquely mapped into a senquece of node and edge additions
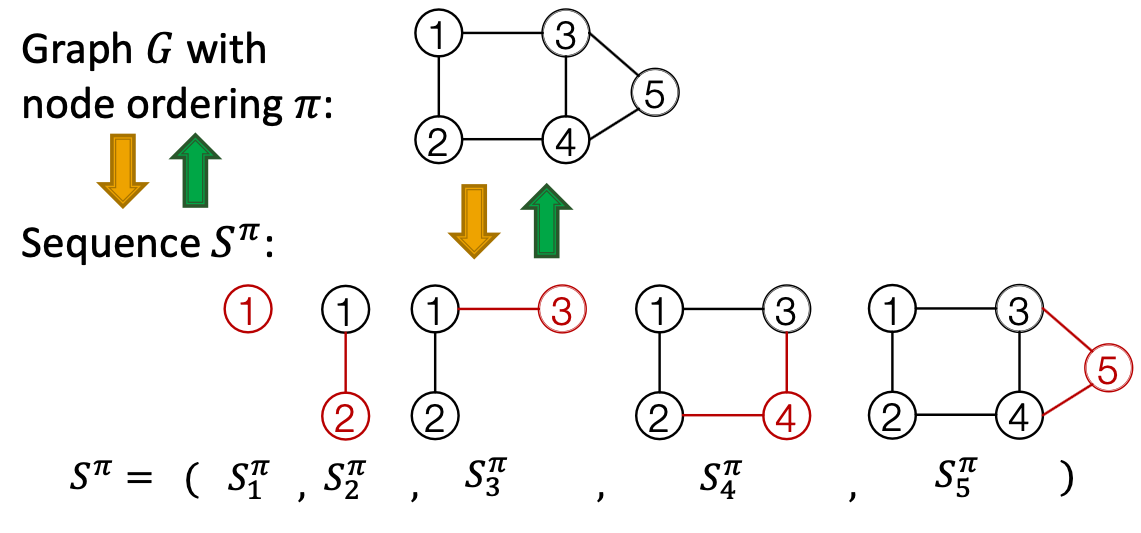
The sequence has two levels ( is a sequence of sequences):
- Node-level: add nodes, one at a time (At each step, a new node is added)
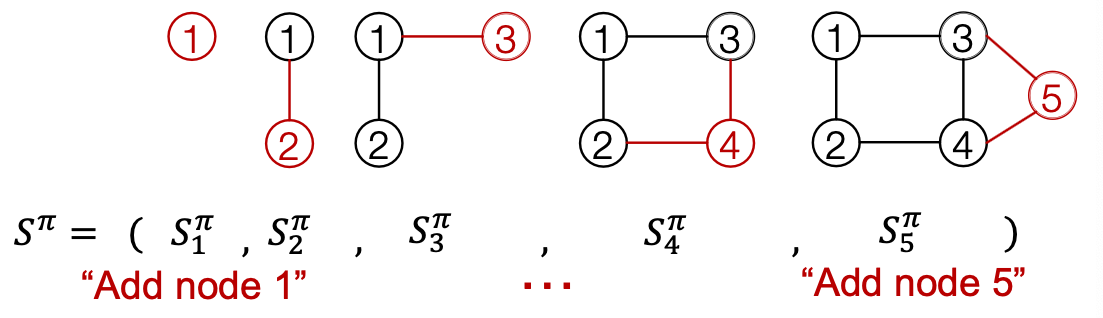
- Edge-level: add edges between existing nodes
- Each Node-level step is an edge-level sequence
- Edge-level: At each step, add a new edge
Summary: A graph + a node ordering = A sequence of sequences
Node ordering is randomly selected (we will come back to this)
We have transformed graph generation problem into a sequence generation problem
Need to model two processes:
- Generate a state for a new node (Node-level sequence)
- Generate edges for the new node based on its state (Edge-level sequence)
Approach: Use Recurrent Neural Networks (RNNs) to model these process!
Background: Recurrent NNs
- RNNs are designed for sequential data
- RNN sequentially takes input sequence to update its hidden states
- The hidden states summarize all the information input to RNN
- The update is conducted via RNN cells
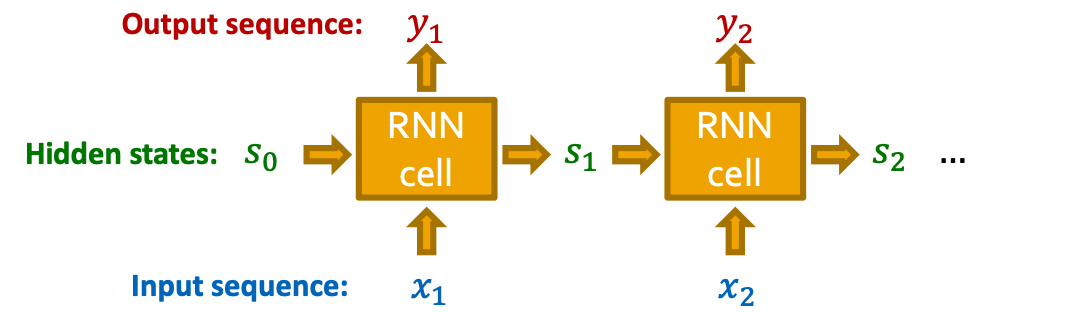
- : State of RNN after step
- : Input to RNN at step
- : Output of RNN at step
- RNN cell: : Trainable parameters
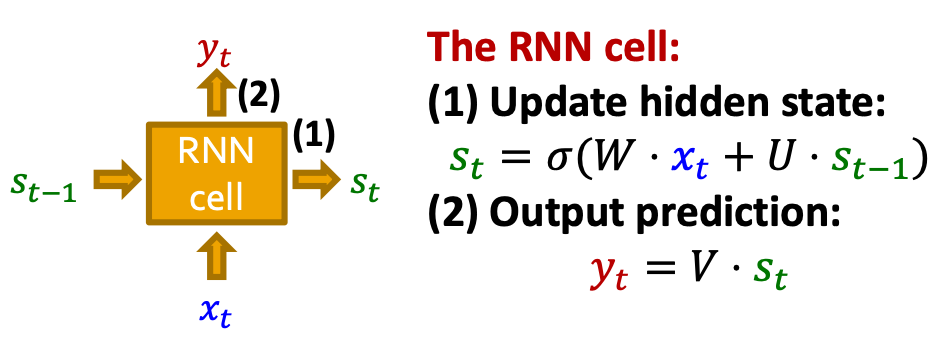
- More expressive cells: GRU, LSTM, etc.
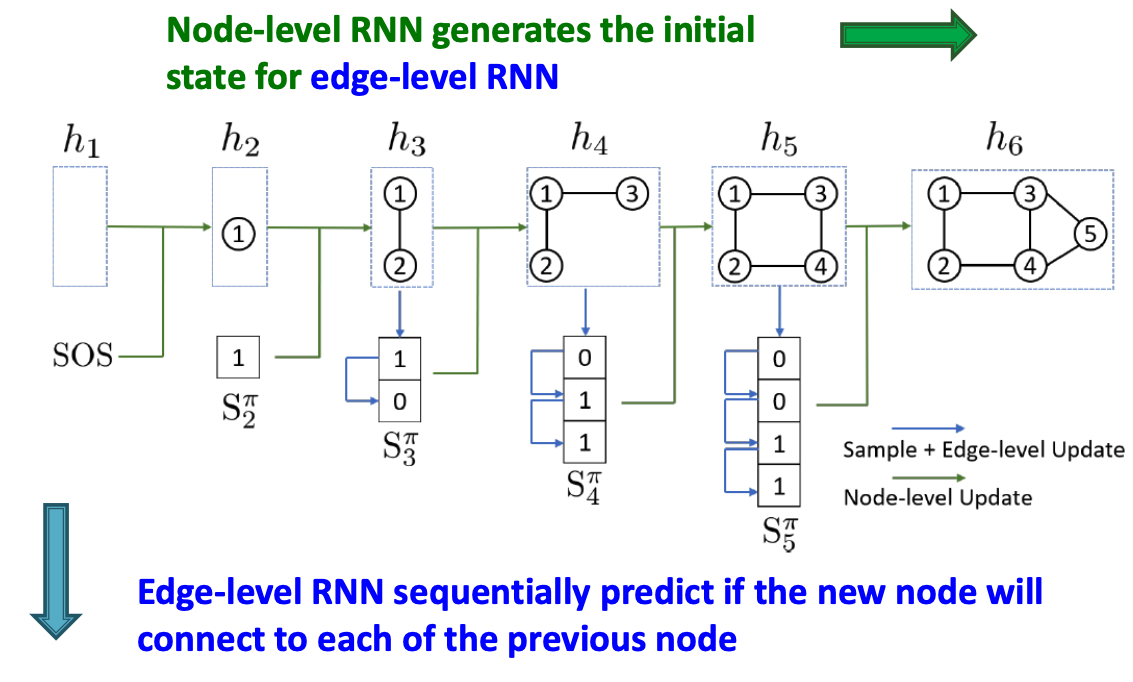
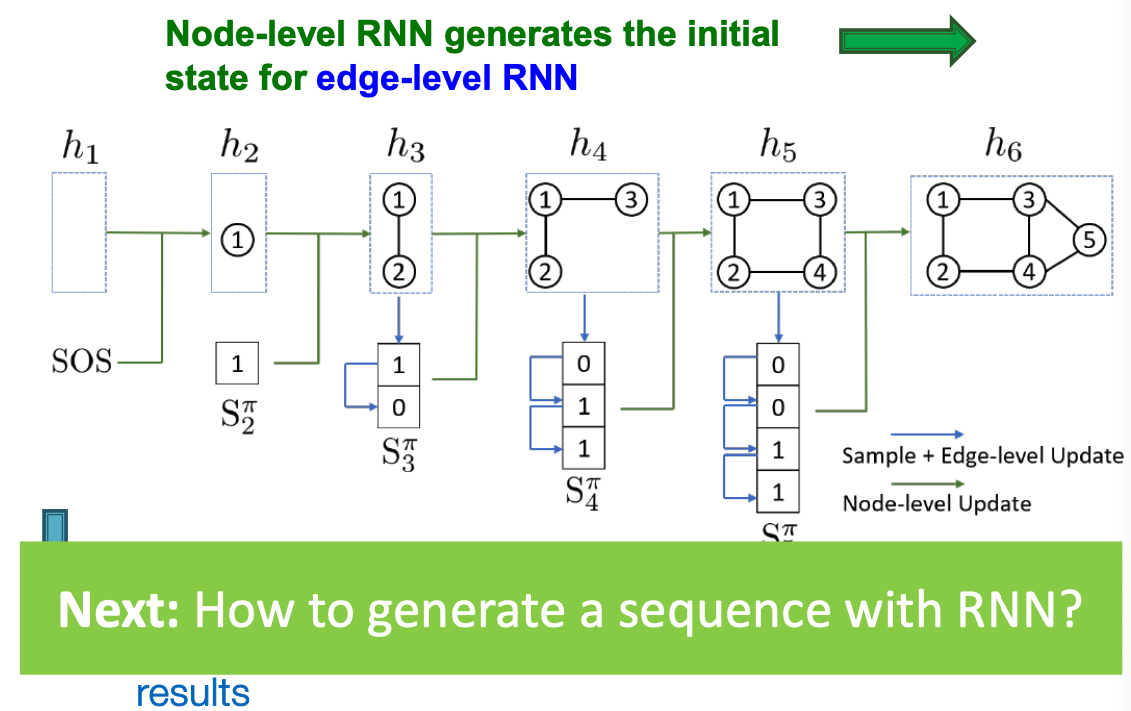
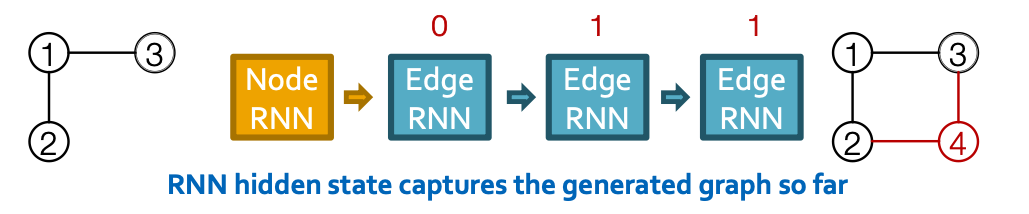
GraphRNN: Two levels of RNN
- GraphRNN has a node-level RNN and an edge-level RNN
- Relationship between the two RNNs:
- Node-level RNN generations the initial state for edge-level RNN
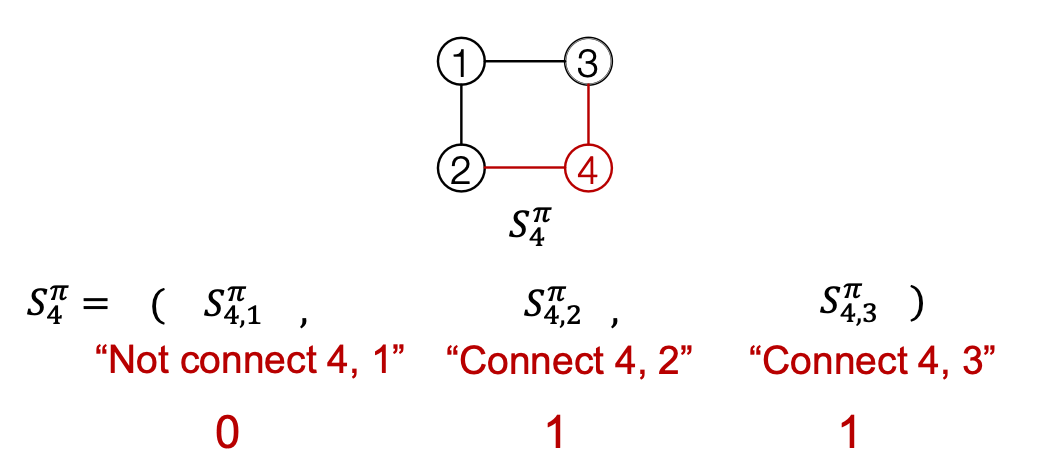
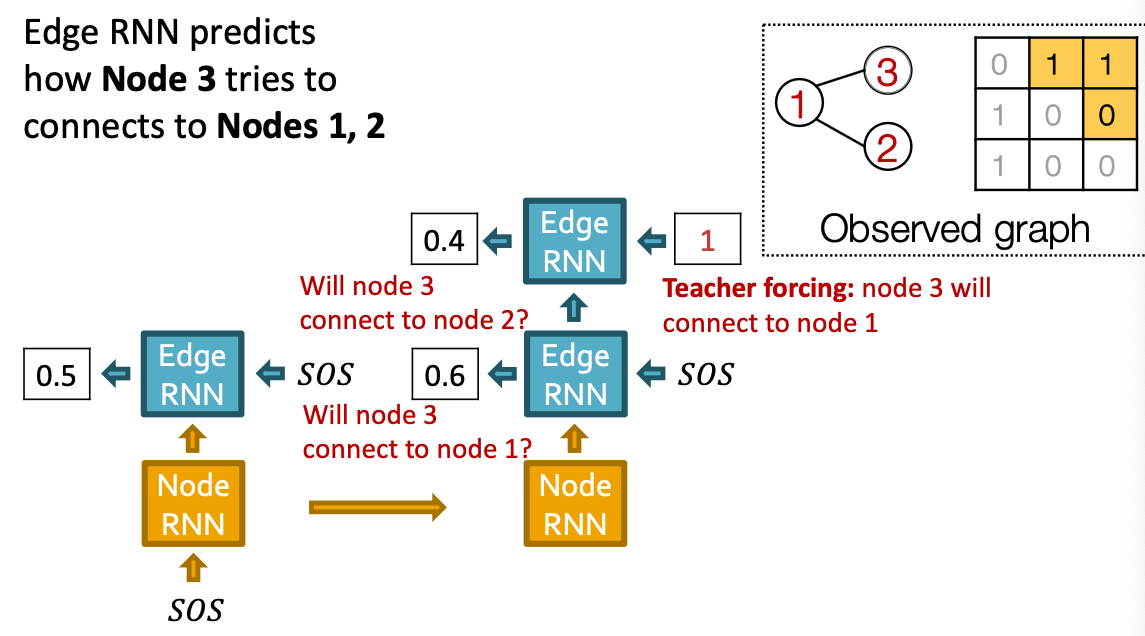
- Edge-level RNN sequentially predict if the new node will connect to each of the previous node.
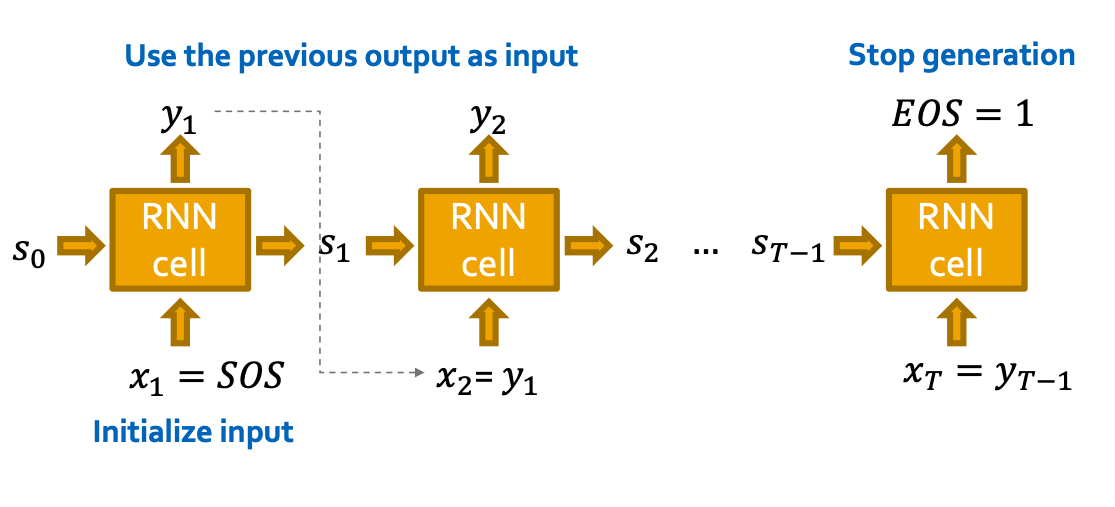
RNN for Sequence Generation
- How to use RNN to generate sequences?
- Let (Use the previous output as input)
- How to initialize the input sequence?
- Use start of sequence token (SOS) as the initial input
- SOS is usually a vector with all zero / ones
- When to stop generation?
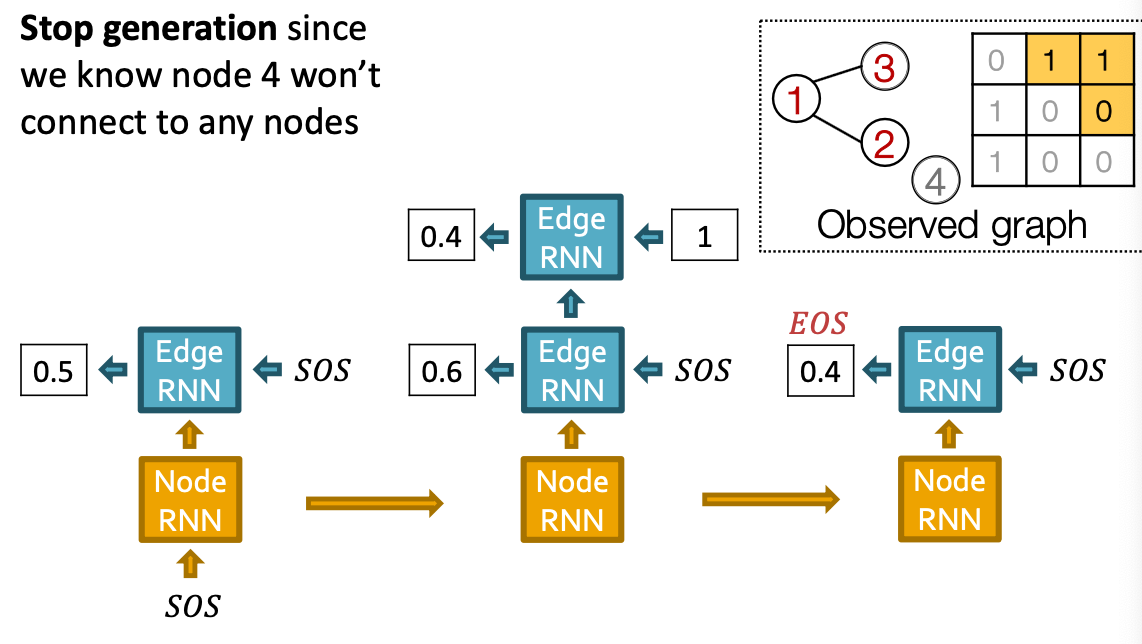
- Use end of sequence token (EOS) as an extra RNN output
- If output EOS=0, RNN will continue generation
- If output EOS =1, RNN will stop generation.
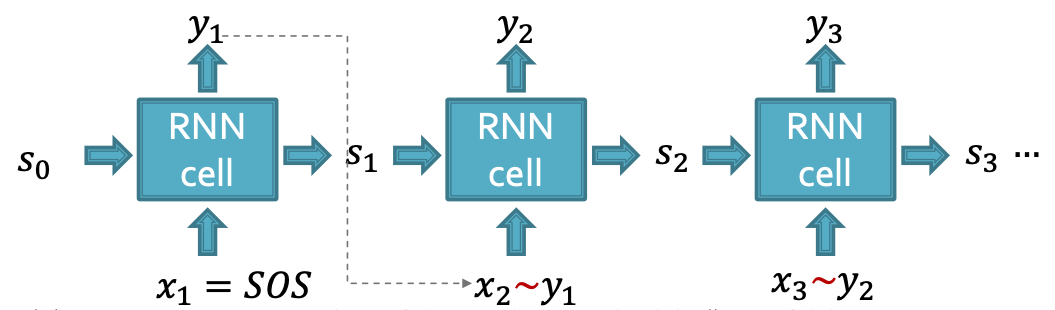
Towards Edge-Level RNN
Consider the Edge-level RNN for now.
- Out goal: Mode
- Let
- The we need to sample from
- Each step of RNN outputs a probability of a single edge
- We then sample from the distribution, and feed sample to next step:
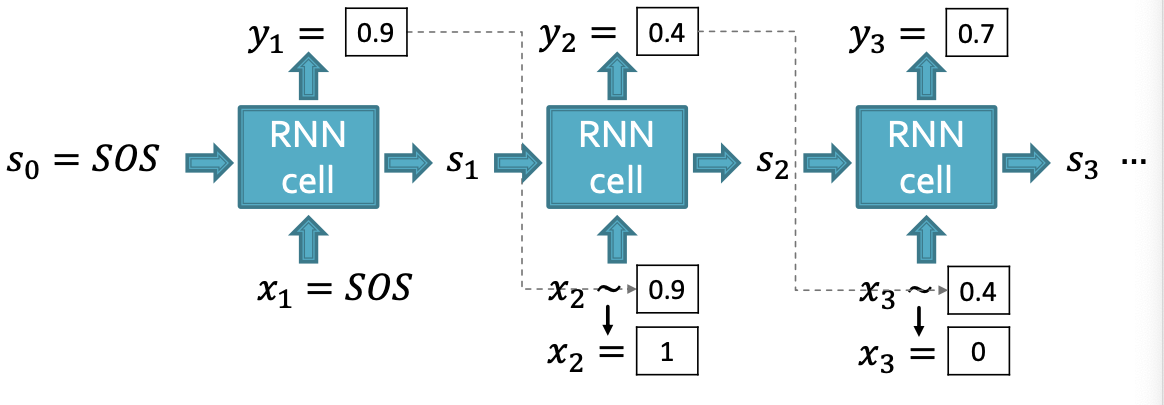
Suppose we already have trained the edge-level RNN
- is a scalar, following a Bernoulli distribution
- means values 1 has probability , value 0 has probability
Edge-level RNN at Training Time
Training the model:
- We observe a sequence of edges
- Principle: Teacher Forcing - Replace input and output by the real sequence
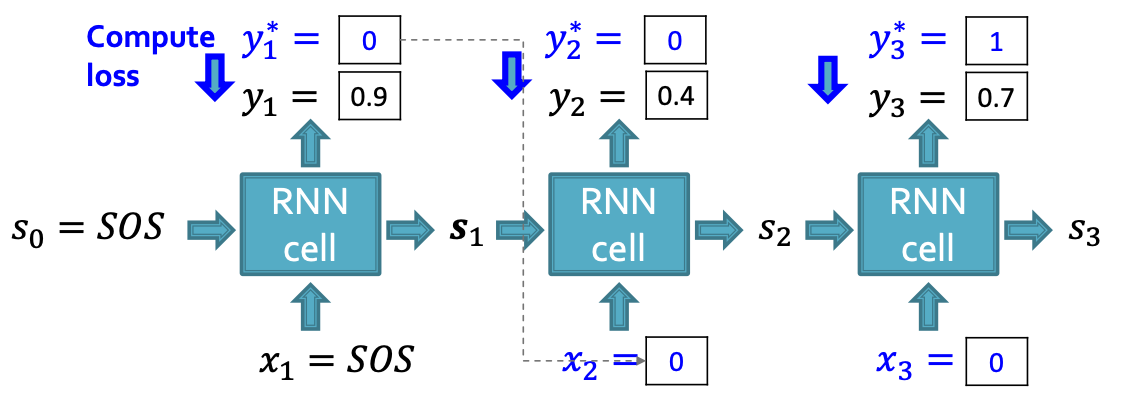
- Loss : Binary cross entropy
- Minimize:
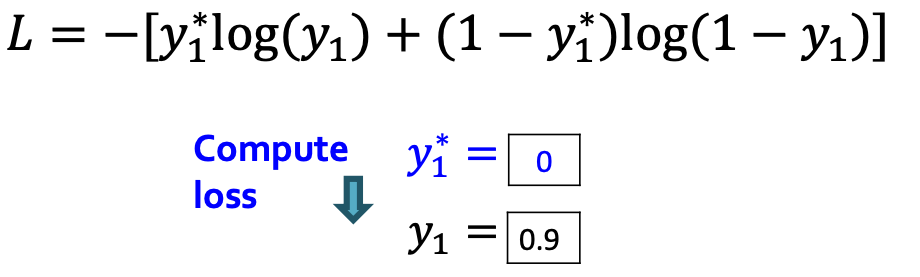
- If , we minimize ,making higher
- If , we minimize , making lower.
- This way, is fitting the data samples
- Reminder: is computed by RNN, this loss will adjust RNN parameters accordingly, using back propagation!
Putting Things Together
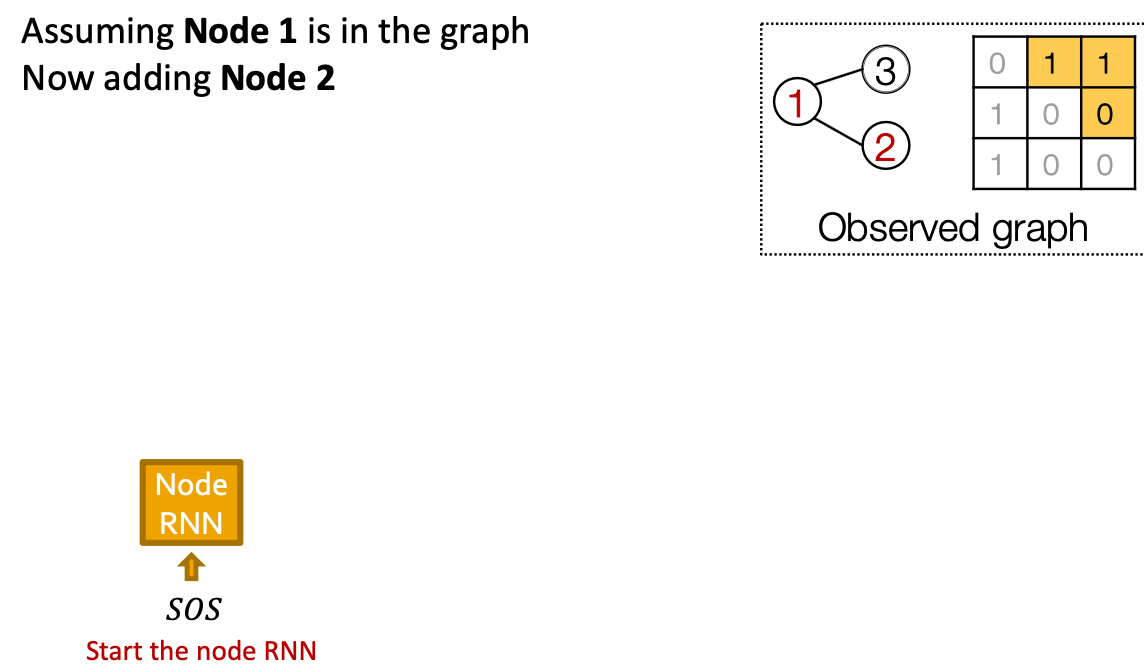
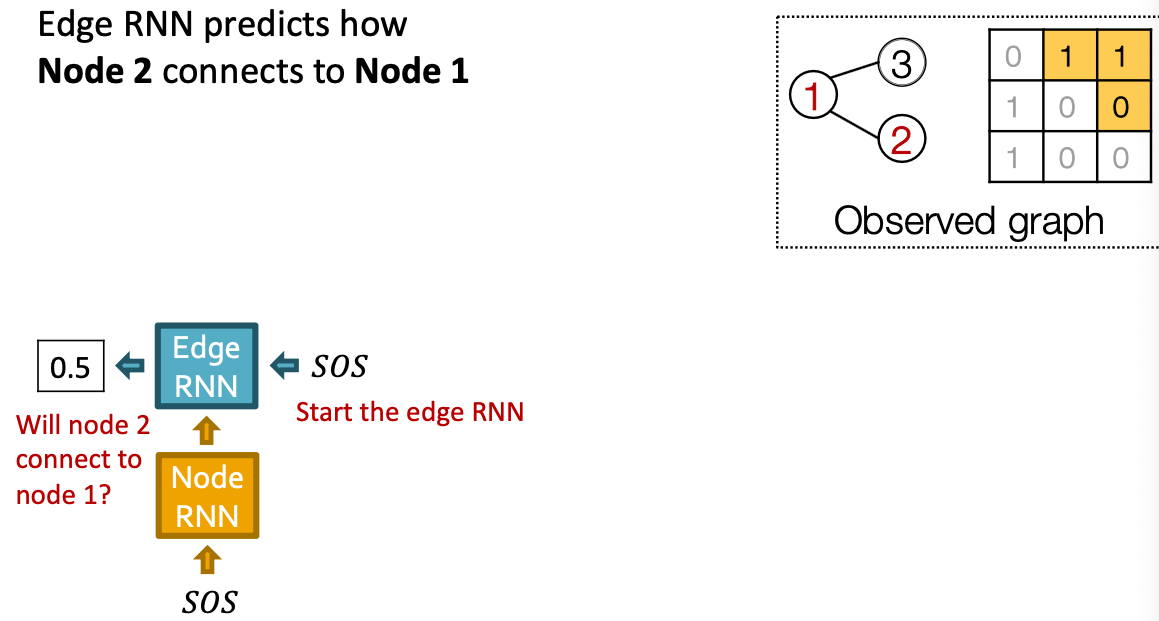
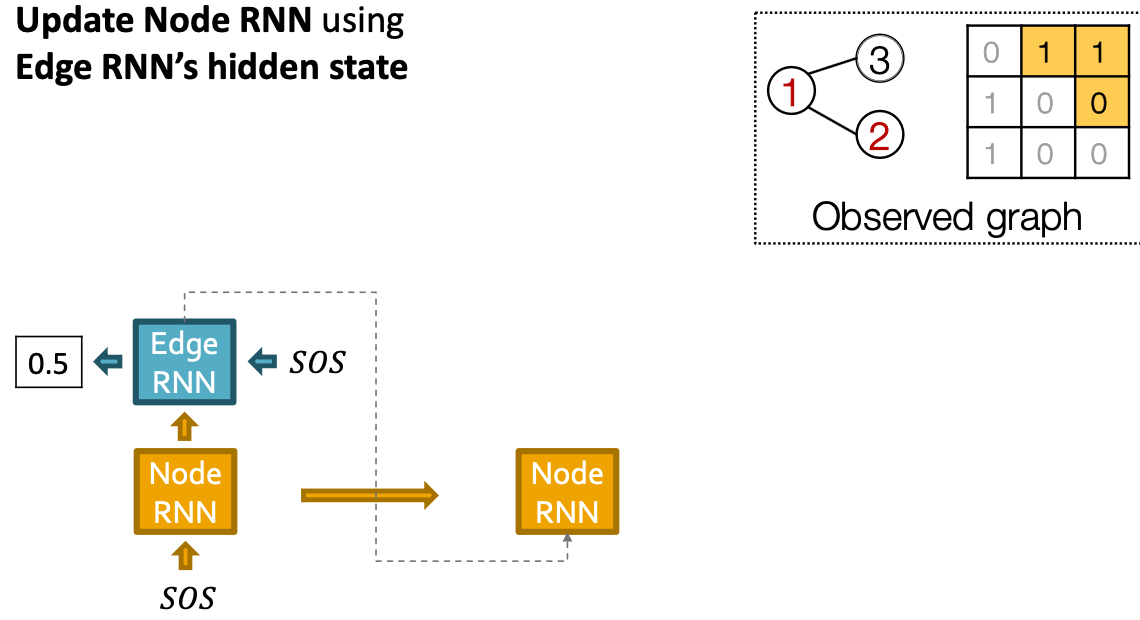
Our Plan:
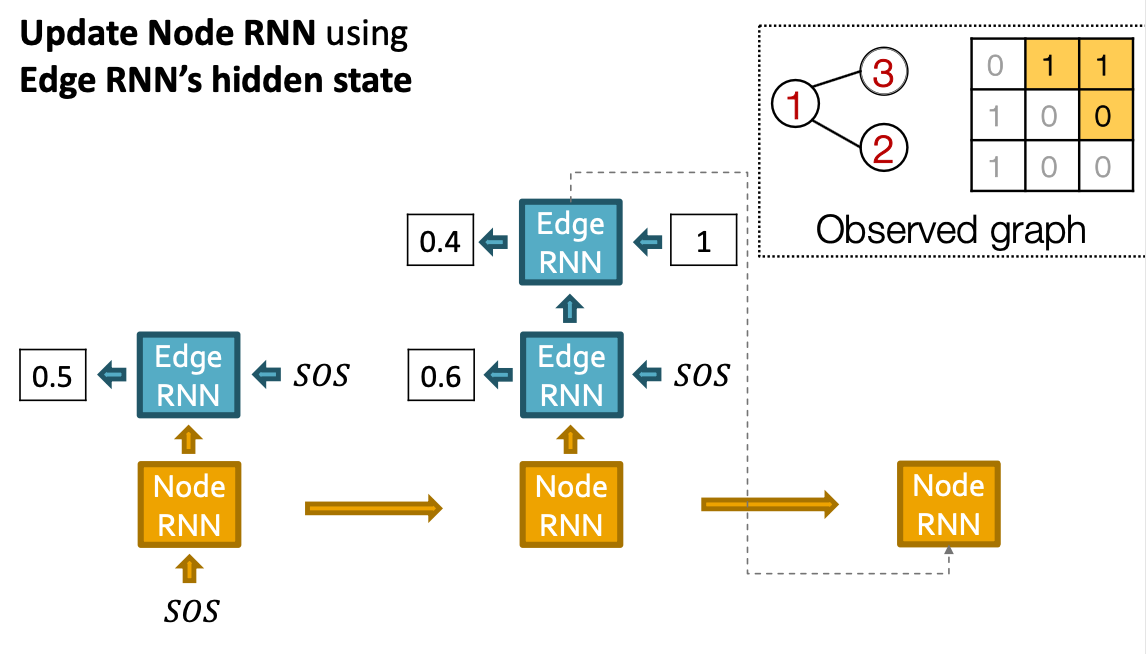
- Add a new node: We run Node RNN for a step, and use it output to initialize Edge RNN.
- Add new edges for the new node: We run Edge RNN to predict if the new node will connect to each of the previous node.
- Add another new node: We use the last hidden state of Edge RNN to run Node RNN for another step
- Stop graph generation: If Edge RNN outputs EOS at step 1, we know no edges are connected to the new node. We stop the graph generation.
Put Things Together: Training
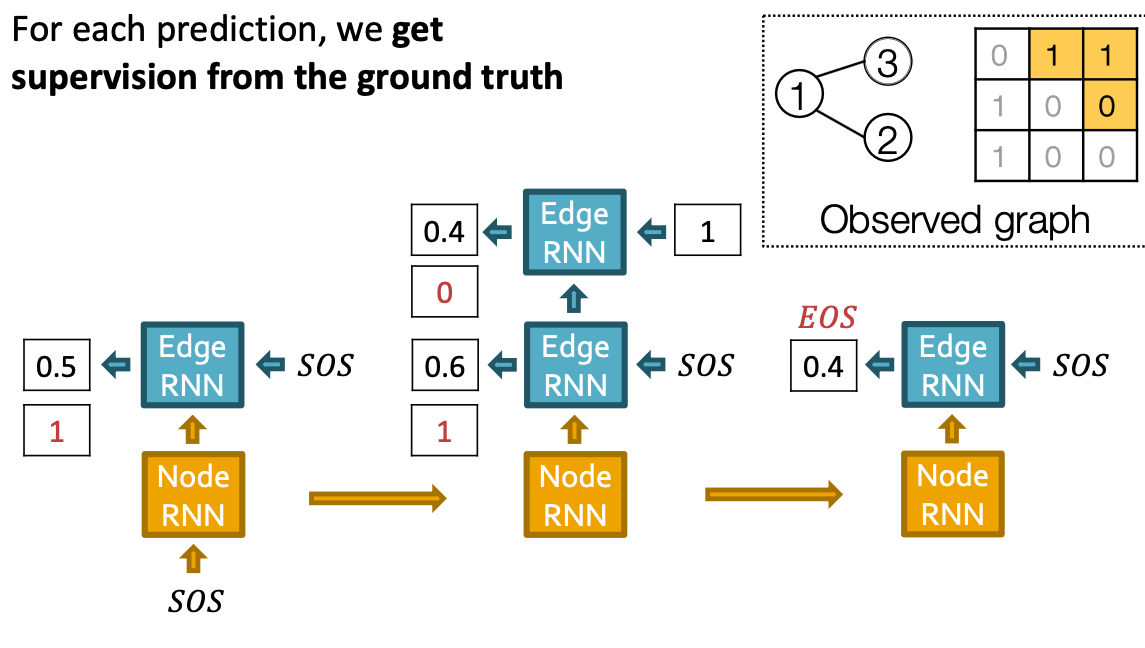
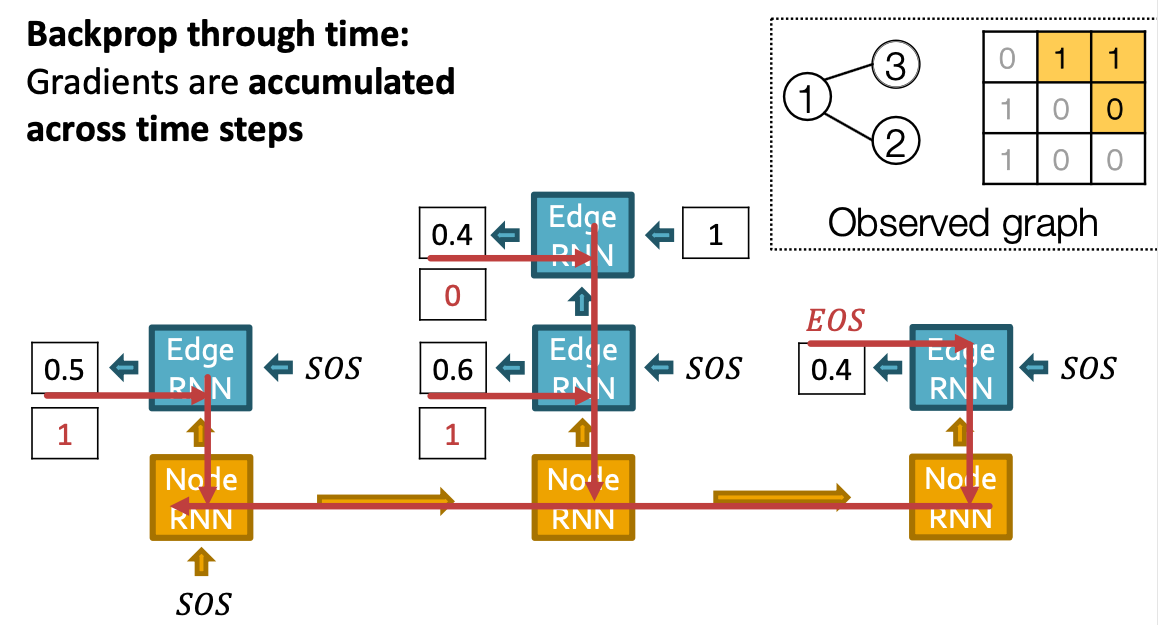
Put Things Together: Test
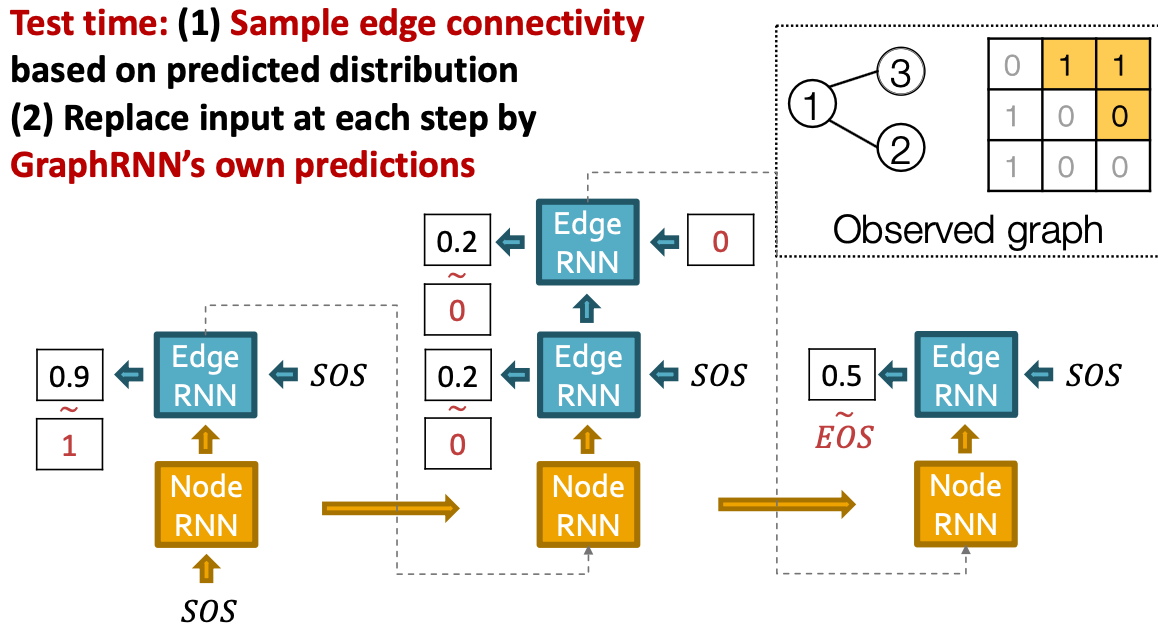
Scaling Up and Evaluating Graph Generation
Issue: Tractability
- Any node can connect to any prior node.
- Too many steps for edge generation
- Need to generate full adjacency matrix
- Complex too-long edge dependencies

Solution: Tractability via BFS
- Breadth-First Search node ordering
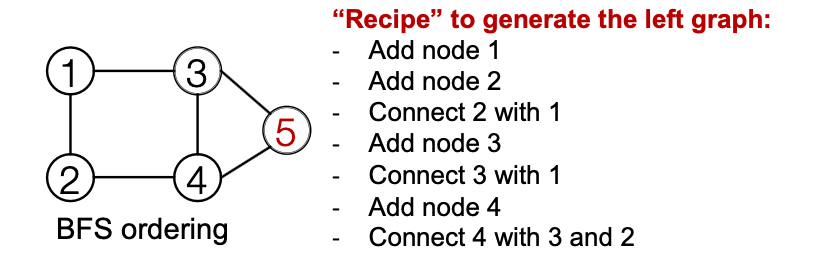
- BFS node ordering:
- Since Node 4 doesn’t connect to Node 1
- We know all Node 1’s neighbors have already been traversed
- Therefore, Node 5 and the following nodes will never connect to node 1
- We only need memory of 2 “steps” rather than steps
- Breadth-First Search ordering

- Benefits:
- Reduce possible node orderings
- From to number of distinct BFS orderings
- Reduce steps for edge generation
- Reducing number of previous nodes to look at
- Reduce possible node orderings
- BFS reduces the number of steps for edge generation
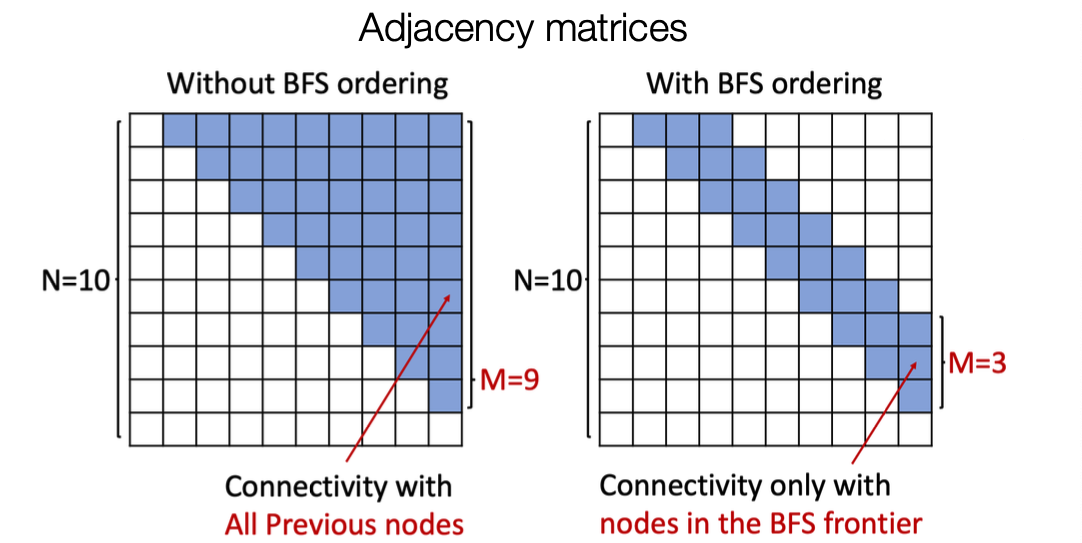
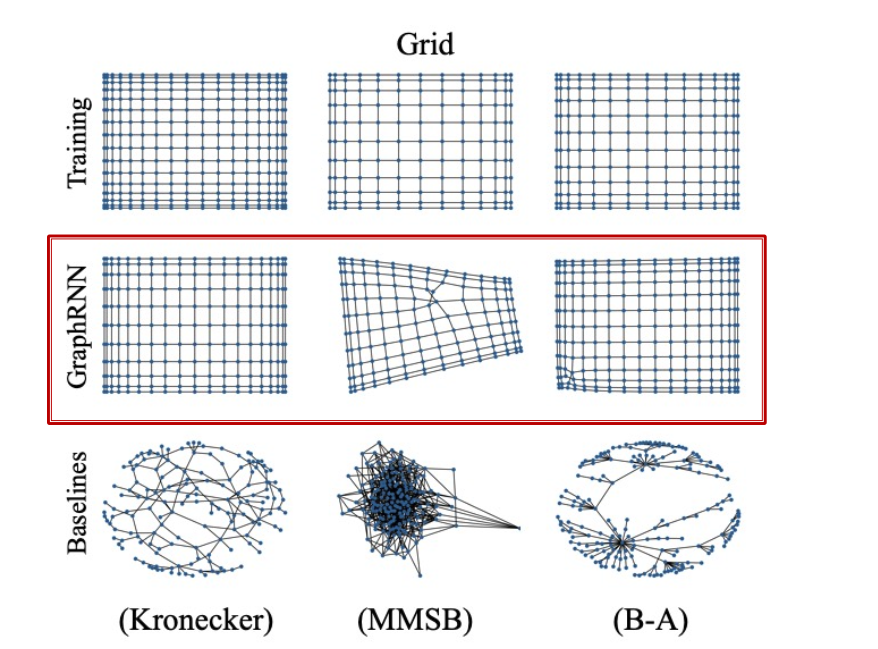
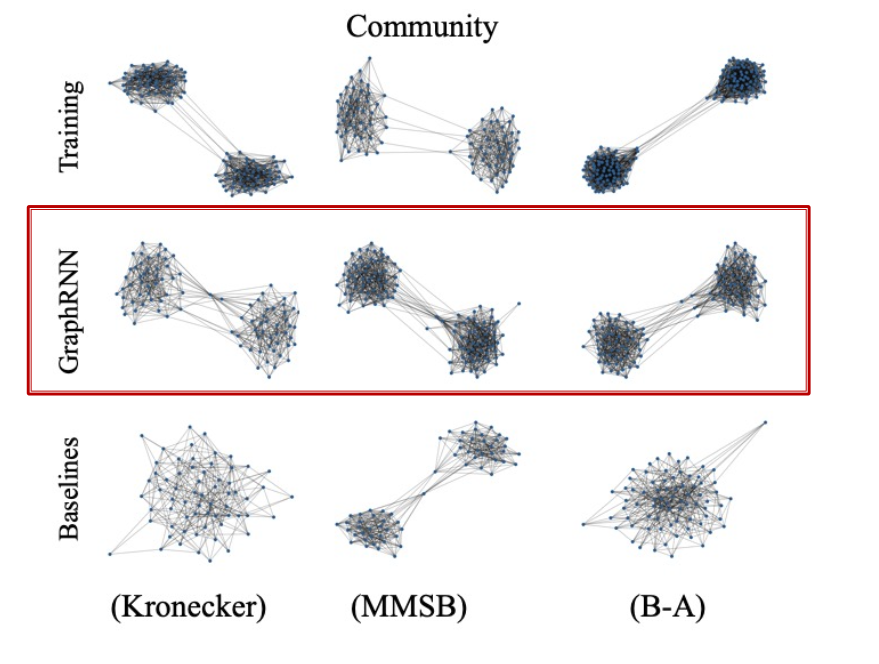
Evaluating Generated Graphs
- Task: Compare two sets of graphs
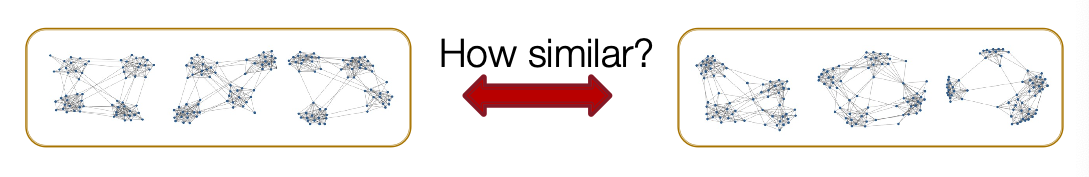
- Goal: Define similarity metrics for graphs
Application of Deep Graph Generative Models to Molecule Generation
Application: Drug Discovery
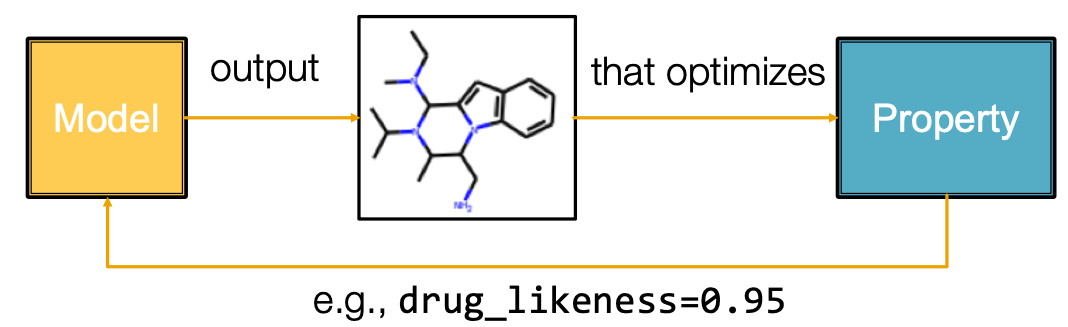
Goal-Directed Graph Generation
Generating graphs that:
- Optimize a given objective (High scores) - Idea: Reinforcement Learning
- E.g., drug-likeness
- Obey underlying rules (Valid)
- E.g., chemical validity rules
- Are learned from examples (Realistic)
- Imitating a molecule graph dataset.
- We have just covered this part.
- Imitating a molecule graph dataset.
Solution: GCPN
Graph Convolutional Policy Network (GCPN)
combines graph representation + RL
Key component of GCPN:
- Graph Neural Network captures graph structural information
- Reinforcement Learning guides the generation towards the desired objectives
- Supervised training imitates examples in given datasets
GCPN vs. GraphRNN
- Commonality of GCPN & GraphRNN
- Generate graphs sequentially
- Imitate a given graph dataset
- Main Differences:
- GCPN uses GNN to predict the generation action
- Pros: GNN is more expressive than RNN
- Cons: GNN takes longer time to compute than RNN
- GCPN further uses RL to direct graph generation to our goals
- RL enables goal-directed graph generation
- GCPN uses GNN to predict the generation action
- Sequential graph generation
- GraphRNN: predict action based on RNN hidden states
- GCPN: predict action based on GNN node embeddings
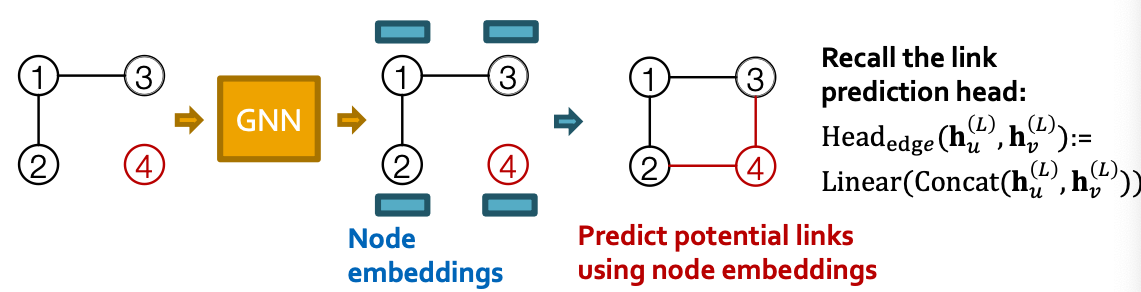
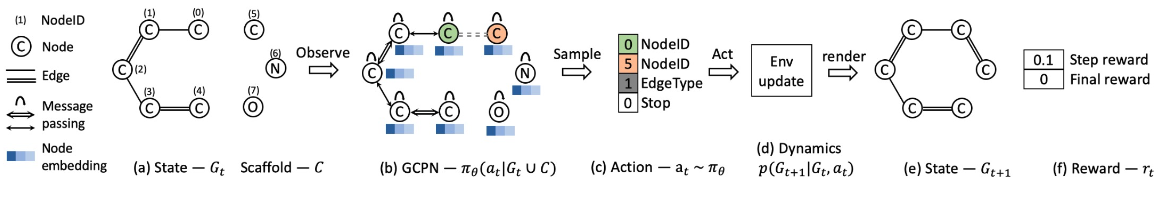
Overview of GCPN
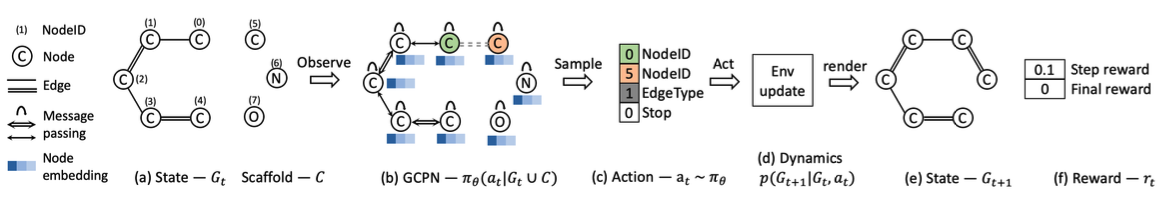
- (a) Insert nodes
- (b c) Use GNN to predict which nodes to connect
- (d) Take an action (check chemical validity)
- (e f) Compute reward
How to set the Reward?
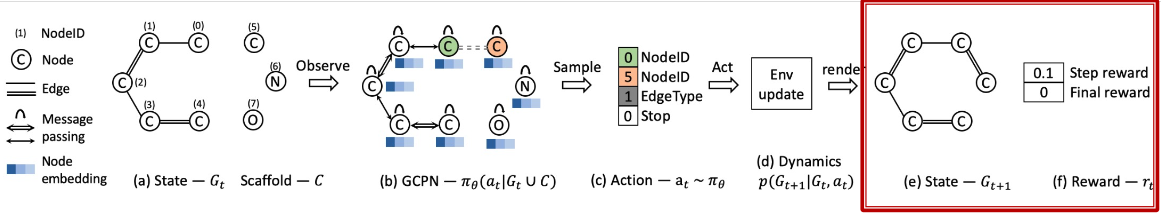
- Step reward: Learn to take valid action
- At each step, assign small positive reward for valid action
- Final reward: Optimize desired properties
- At the end, assign positive reward for high desired property
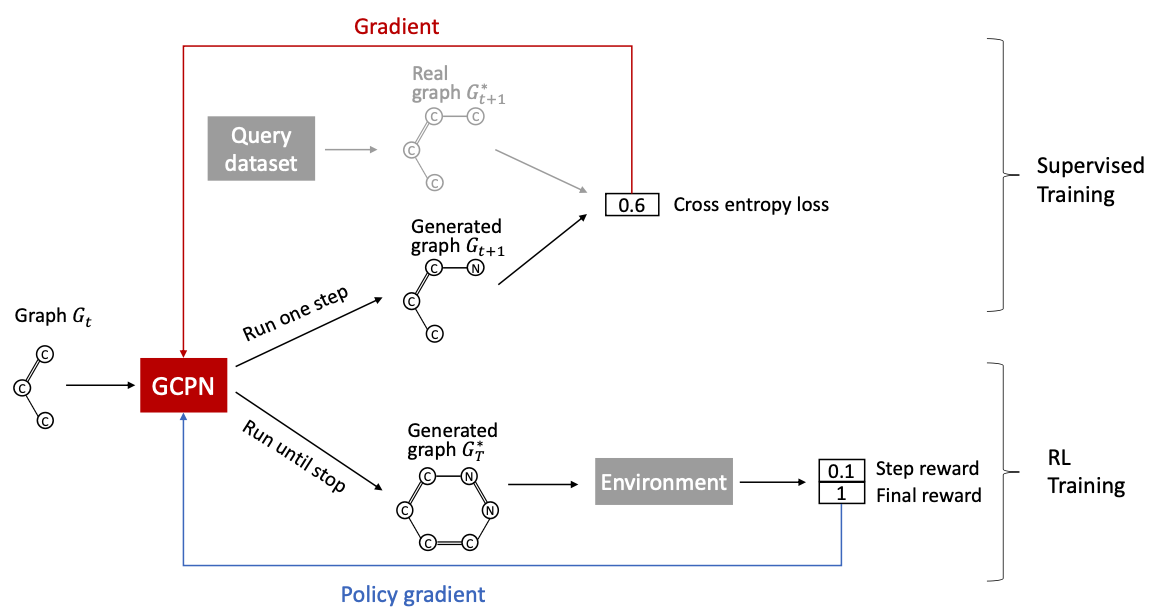
How to train?
- Supervised training: Train policy by imitating the action given by real observed graphs. use gradient.
- We have covered this idea in GraphRNN
- Reinforcement Learning training: Train policy to optimize rewards. Use standard policy gradient algorithm.
Training GCPN
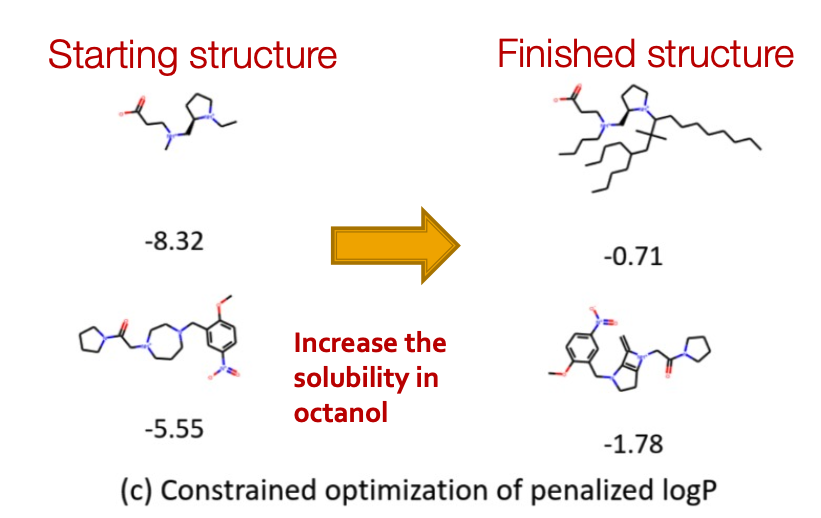
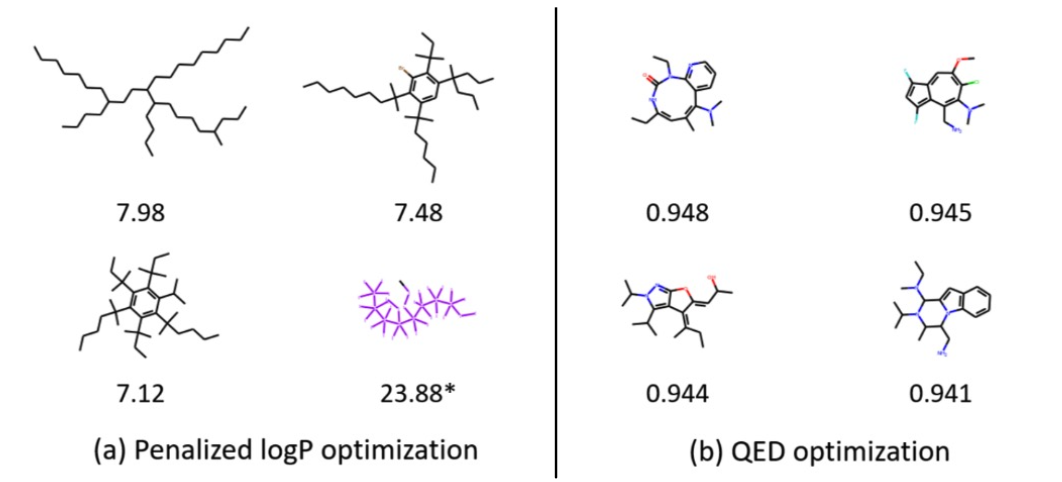
Qualitative Results
Visualization of GCPN graphs:
- Property optimizattion Generate molecules with high specified property score
- Constrained Optimization: Edit a given molecule for a few steps to achieve higher property score