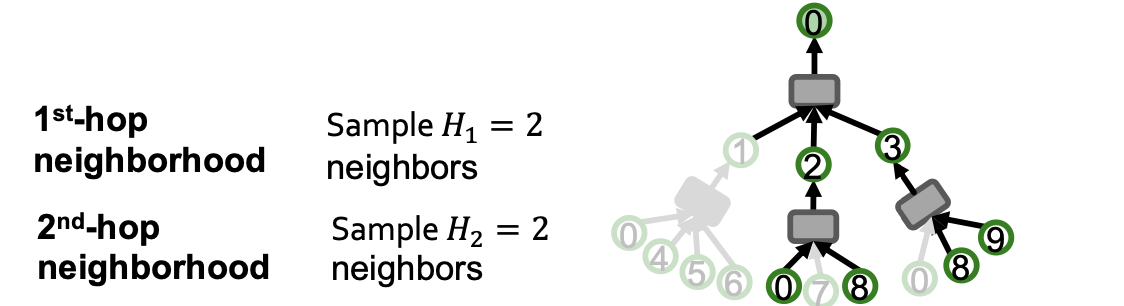CS224W-Machine Learning with Graph-Scaling Up GNNs
- Large - Scale:
- #nodes ranges from 10M to 10B
- #edges ranges from 100M to 100B
- Tasks
- Node-level: User/item/paper classification
- Link-level: Recommendation, completion
Standard SGD cannot effectively train GNNs
- Objective: Minimize the average loss
- : model parameters
- : loss for -th data point
- We perform Stochastic Gradient Descent (SGD)
- Sample data points (mini-batches).
- Compute the over the data points.
- Perform SGD:
In mini-batch, we sample nodes independently:
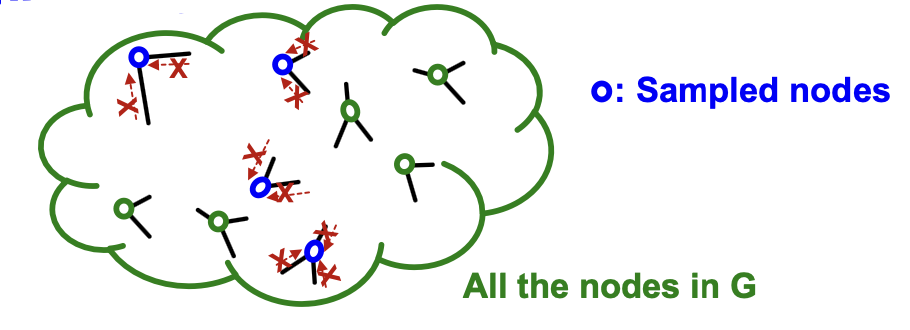
- Sampled nodes will be isolated from each other!
- GNN generates node embeddings by aggregating neighboring node features.
- GNN does not access to neighboring nodes within the mini-batch!
Naive full-batch implementation: Generate embeddings of all the nodes at the same time:
- Load the entire graph and features . Set
- At each GNN layer: Compute embeddings of all nodes using all the node embeddings from the previous layer.
- Compute the loss
- Perform gradient descent
Full-batch implementation is not feasible for a large graphs. → Because we want to use GPU for fast training, but GPU memory is extremely limited (10GB - 80GB). The entire graph and the features can not be loaded on GPU.
Two methods perform message-passing over small subgraph in each mini-batch; only the subgraphs need to be loaded on a GPU at a time.
- Neighbor Sampling [Hamilton et al. NeurIPS 2017]
- Cluster-GCN [Chiang et al. KDD 2019]
One method simplifies a GNN into feature-preprocessing operation (can be efficiently performed even on a CPU)
- Simplified GCN [Wu et al. ICML 2019]
Sampling: Scaling up GNNs
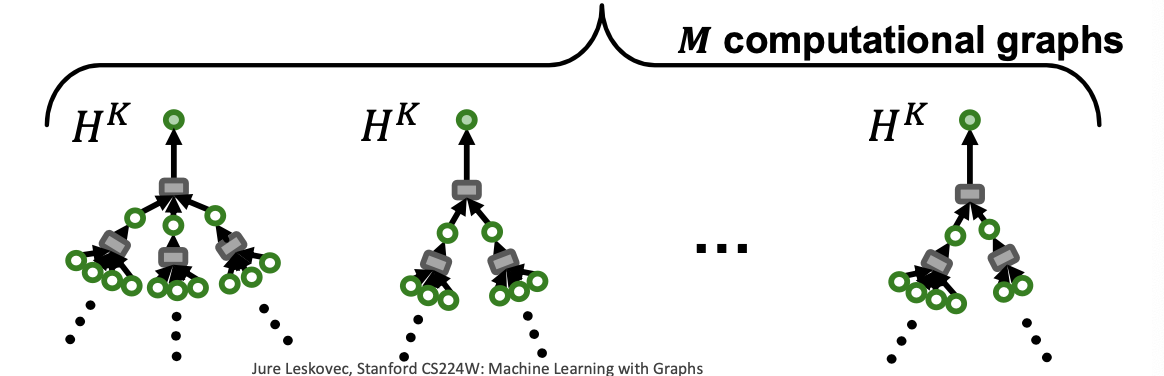
Recall: Computational Graph
- GNNs generate node embeddings via neighbor aggregation
- Represented as a computational graph (right)
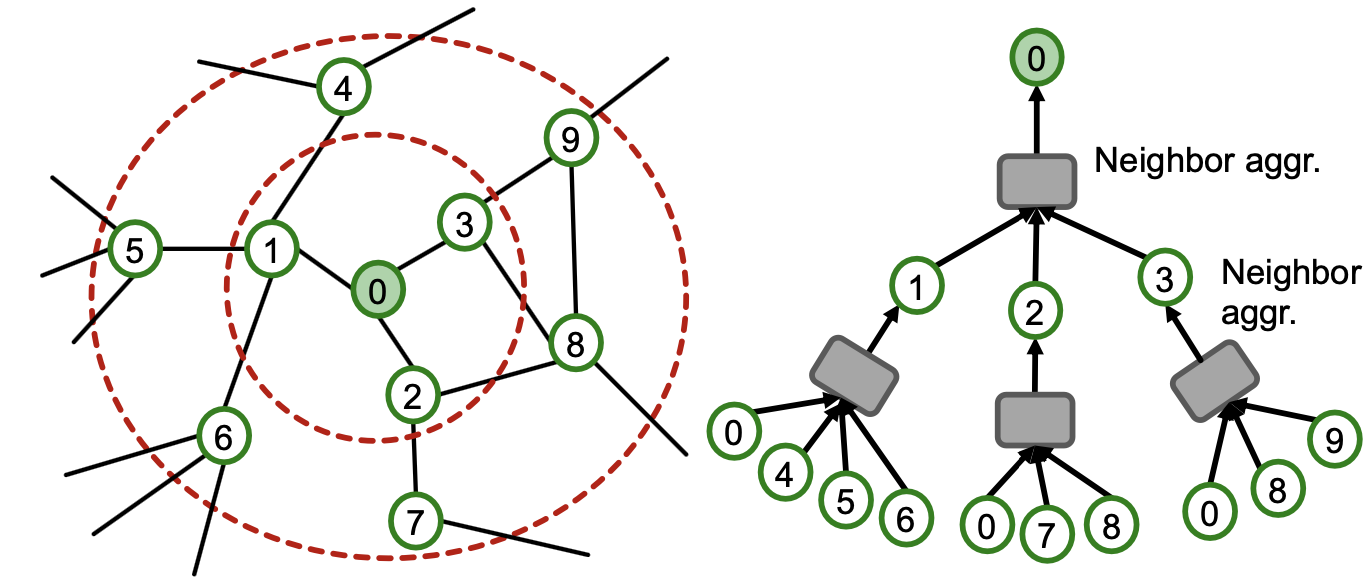
- Observation: A 2-layer GNN generates embedding of node “0” using 2-hop neighborhood structure and features
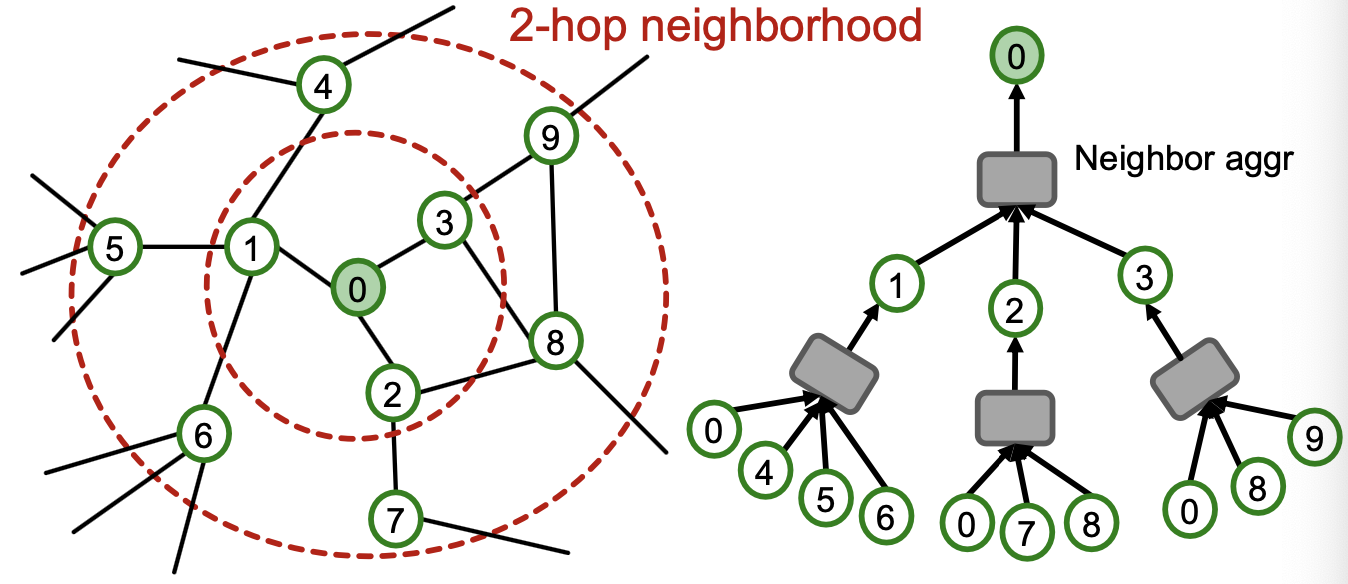
- Observation: More generally, K-layer GNNs generate embedding of a node using -hop neighborhood structure and features.
Computing Node Embeddings
- Key insight: To compute embedding of a single node, all we need is the -hop neighborhood (which defines the computation graph)
- Given a set of different nodes in a mini-batch, we can generate their embeddings using computational graphs. Can be computed on GPU.
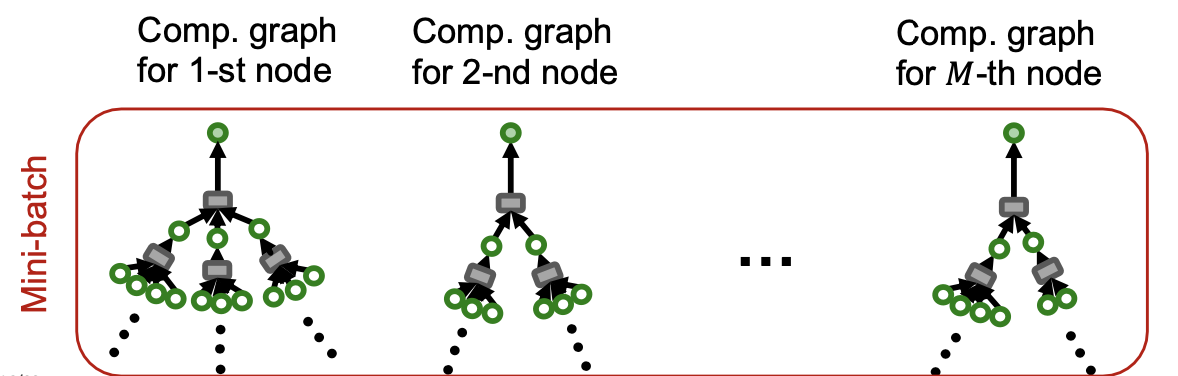
Stochastic Training of GNNs
- We can now consider the following SGD strategy for training -layer GNNs:
- Randomly sample root nodes
- For each sampled root node :
- Get -hop neighborhood and construct the computation graph
- Use the above to generate ’s embedding
- Compute the loss averaged over the nodes
- Perform SGD:
Issue with Stochastic Training
- For each node, we need to get the entire -hop neighborhood and pass it through the computation graph
- We need to aggregate lot of information just to compute one node embedding
- issue:
- Computation graph becomes exponentially large with respect to the layer size .
- Computation graph explodes when it hits a hub node (high-degree node)
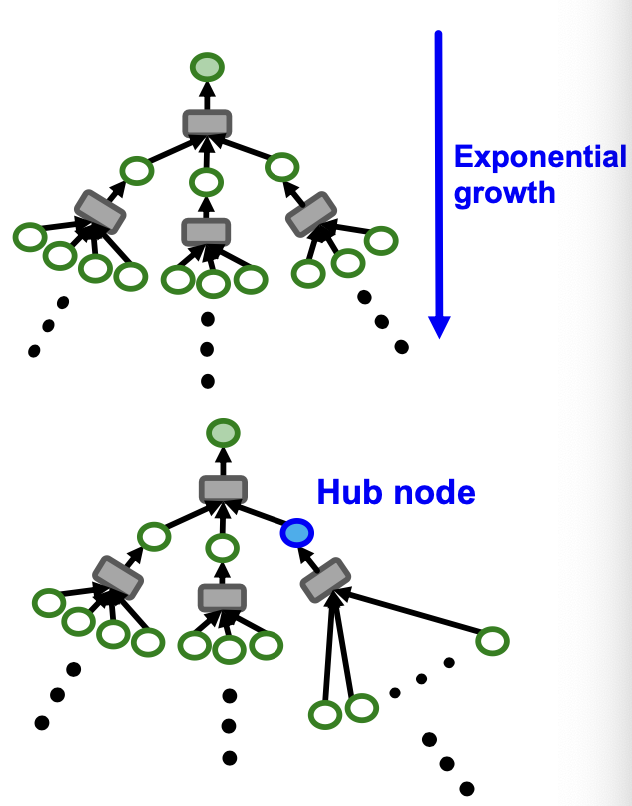
Neighborhood Sampling
Key idea: Construct the computational graph by (randomly) sampling at most neighbors at each hop.
- Example :
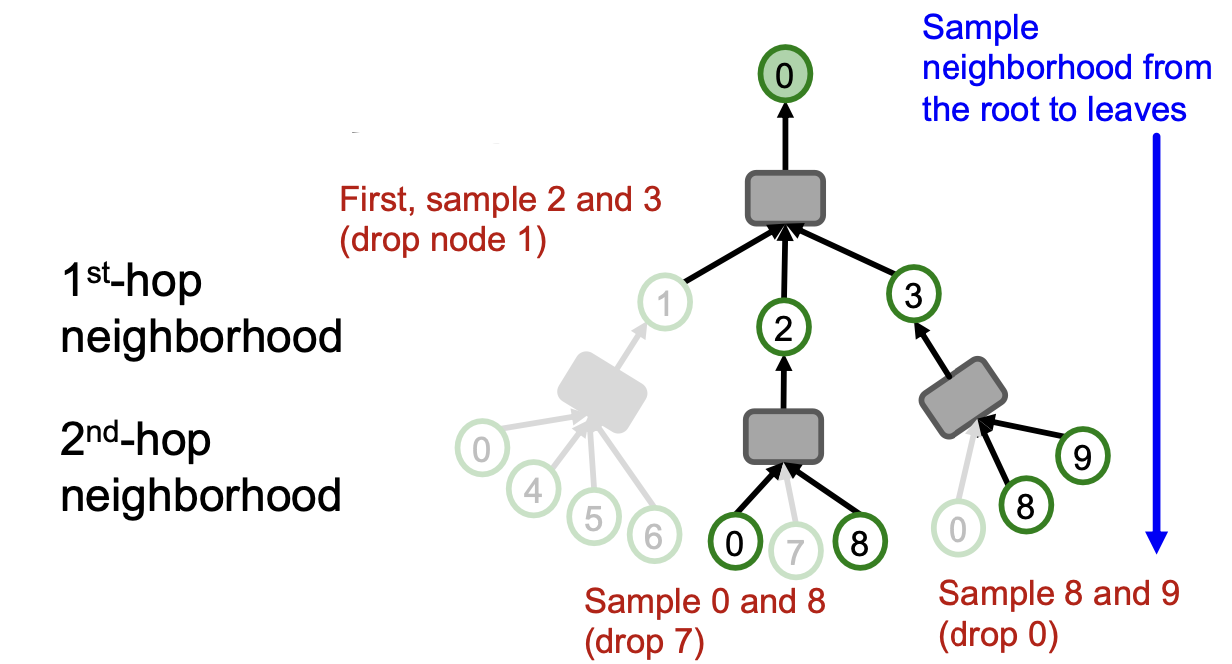
We can use the pruned computational graph to more efficiently compute node embeddings.(剪枝操作)
Neighborhood Sampling Algorithm
Neighbor sampling for -layer GNN
- -layer GNN will at most involve leaf nodes in computational graph
Remarks on Neighbor Sampling
- Remark 1: Trade-off in sampling number
- Smaller leads to more efficient neighbor aggregation, but results are less stable traning due to the large variance in neighbor aggregation
- Remark 2: Computational time
- Even with neighbor sampling, the size of the computational graph is still exponential with respect to number of GNN layers .
- Adding one GNN layer would make computation times more expensive.
- Remark 3: How to sample the nodes
- Random sampling: fast but many times not optimal (may sample many “unimportant” nodes)
- Random Walk with Restarts:
- Natural graphs are “scale free”, sampling random neighbors, samples many low degree “leaf” nodes.
- Strategy to sample important nodes:
- Compute Random Walk with Restarts score starting at the green node
- At each level sample neighbors with the highest
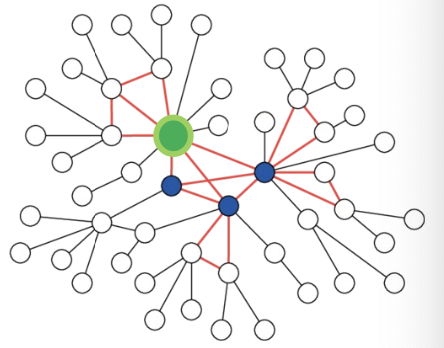
- This strategy works much better in practice.
Scaling Up GNNs
Issues with Neighbor Sampling
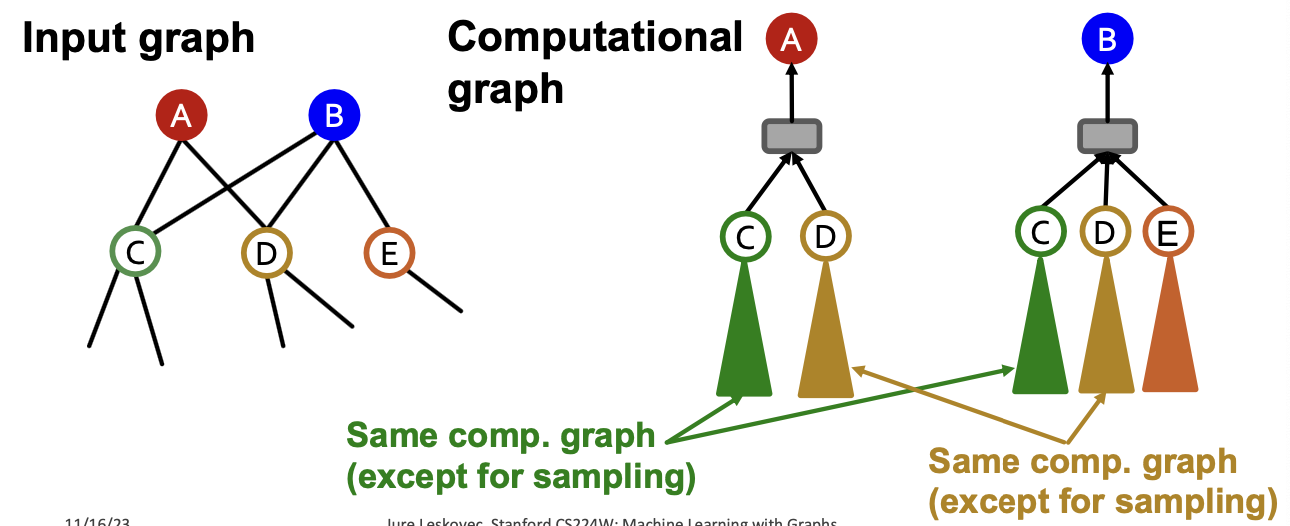
- The size of computational graph becomes exponentially large w.r.t the #GNN layers
- Computation is redundant, especially when nodes in a mini-batch share many neighbors
Recall: Full Batch GNN
- In full-batch GNN implementation, all the node embeddings are updated together using embeddings of the previous layer.
- In each layer, only (edges) messages need to be computed.
- For -layer GNN, only (edges) messages need to be computed
- GNN’s entire computation is only linear in #(edges) and #(GNN layers). Fast!
Update for all
Insight from Full-batch GNN
- The layer-wise node embedding update allows the re-use of embeddings from the previous layer.
- This significantly reduces the computational redundancy of neighbor sampling.
- Of course, the layer-wise update is not feasible for a large graph due to limited GPU memory.
- Requires putting the entire graph and features on GPU.
- Of course, the layer-wise update is not feasible for a large graph due to limited GPU memory.
Subgraph Sampling
- Key idea: We can sample a small subgraph of the large graph and then perform the efficient layer-wise node embeddings update over the subgraph
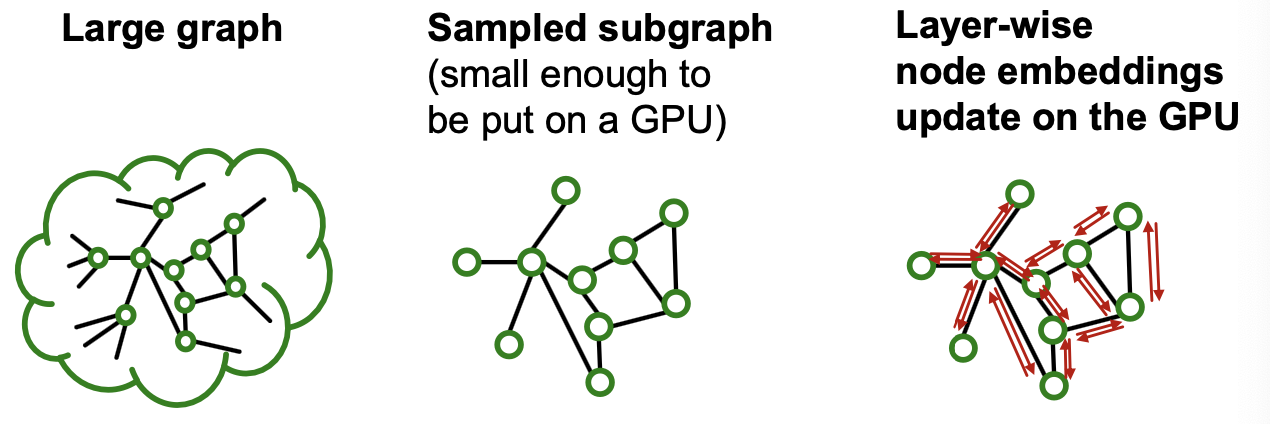
- Key question: What subgraphs are good for training GNNs
- Recall: GNN performs node embedding by passing messages via the edges.
- Subgraphs should retain edge connectivity structure of the original graph as much as possible.
- This way, the GNN over the subgraph generates embeddings closer to the GNN over the original graph.
- Recall: GNN performs node embedding by passing messages via the edges.
Subgraph Sampling: Case Study
- Which subgraph is good for training GNN?
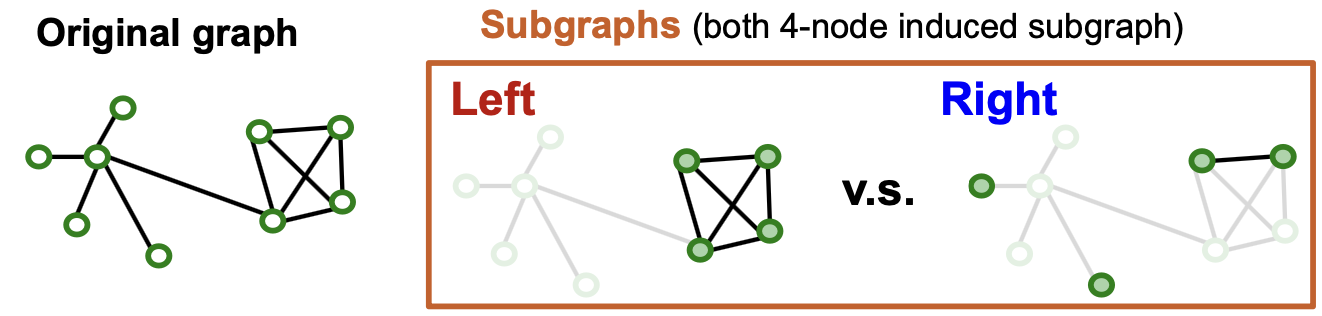
- Left subgraph retrains the essential community structure among the 4 nodes → Good
- Right subgraph drops many connectivity patterns, even leading to isolated nodes → Bad
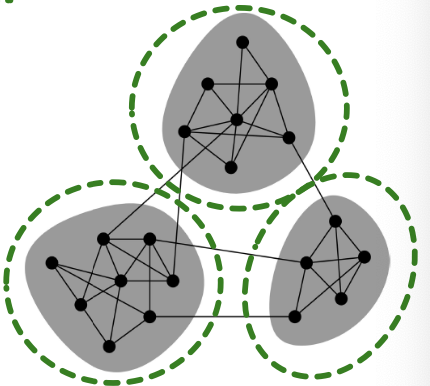
Exploiting Community Structure
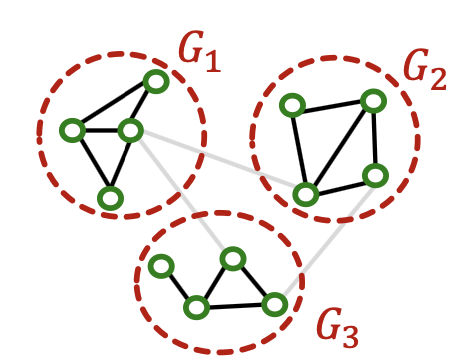
Real-world graph exhibits community structure
- A large graph can be decomposed into many small communities
Key insight: Sample a community as a subgraph. Each subgraph retains essential local connectivity pattern of the original graph.
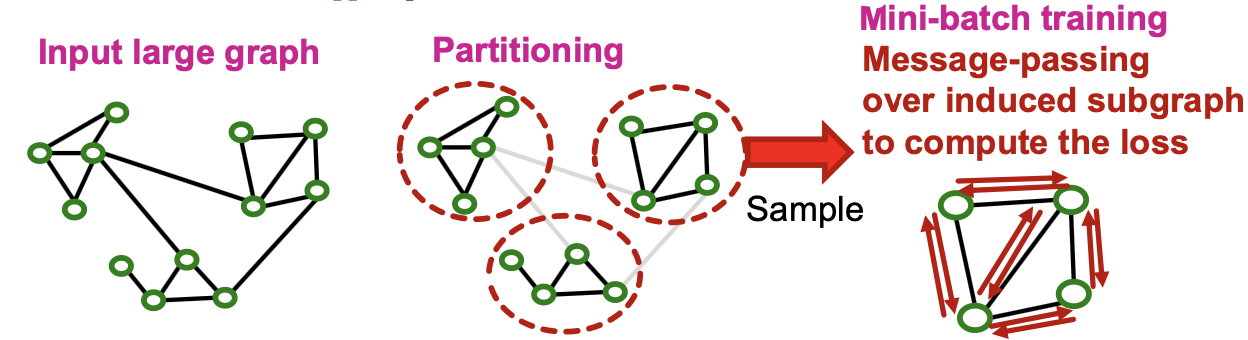
Cluster-GCN: Overview
- We first introduce “vanilla” Cluster-GCN
- Cluster-GCN consists of two steps:
- Pre-processing: Given a large graph, partition it into groups of nodes (i.e., subgraphs).
- Mini-batch training: Sample one node group at a time. Apply GNN’s message passing over the induced subgraph.
Cluster-GCN: Pre-processing
- Given a large graph , partition its nodes into groups:
- We can use any scalable community detection methods, e.g., Louvain, METIS [Karypis et al. SIAM 1998]
Cluster-GCN: Mini-batch Training
- For each mini-batch, randomly sample a node group .
- Construct induced subgraph
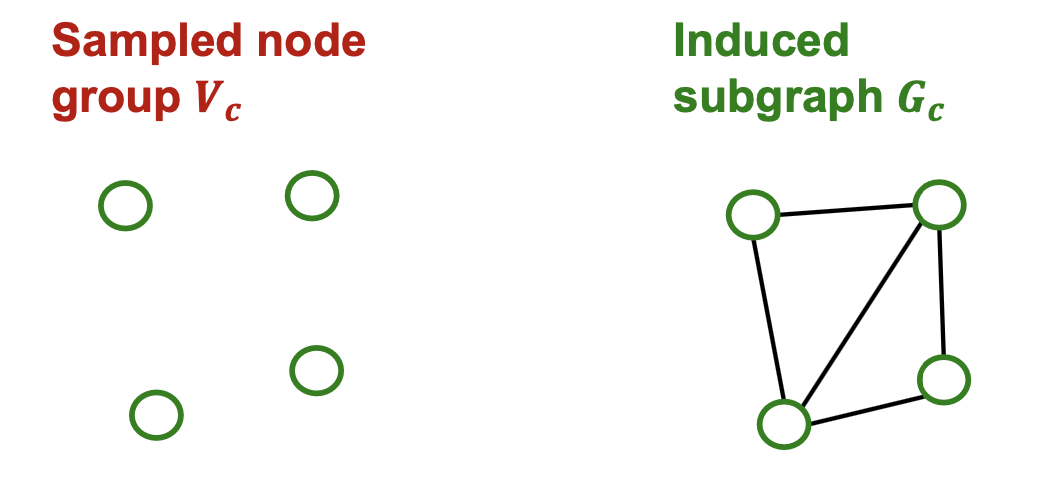
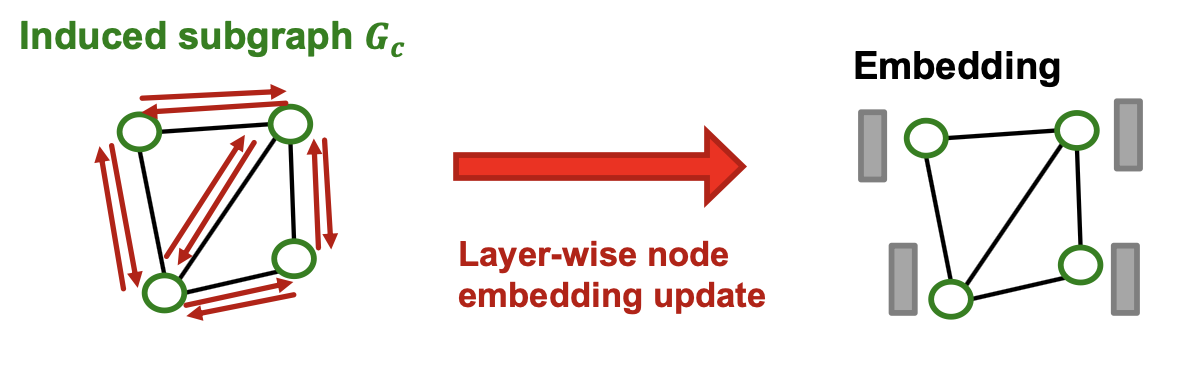
- Apply GNN’s layer-wise node update over to obtain embedding for each node .
- Compute the loss for each node and take average:
- Update params:
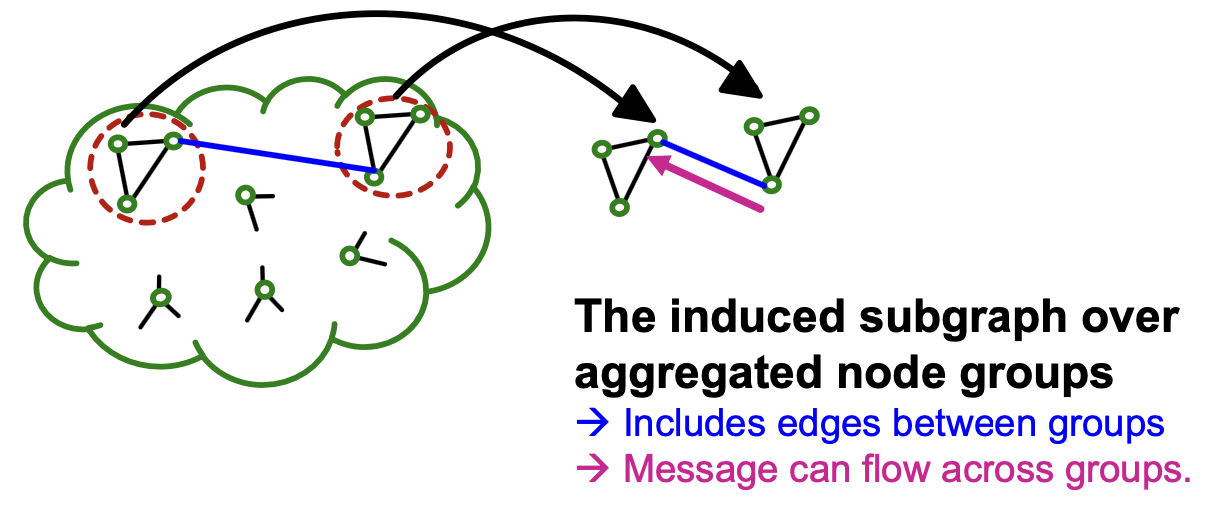
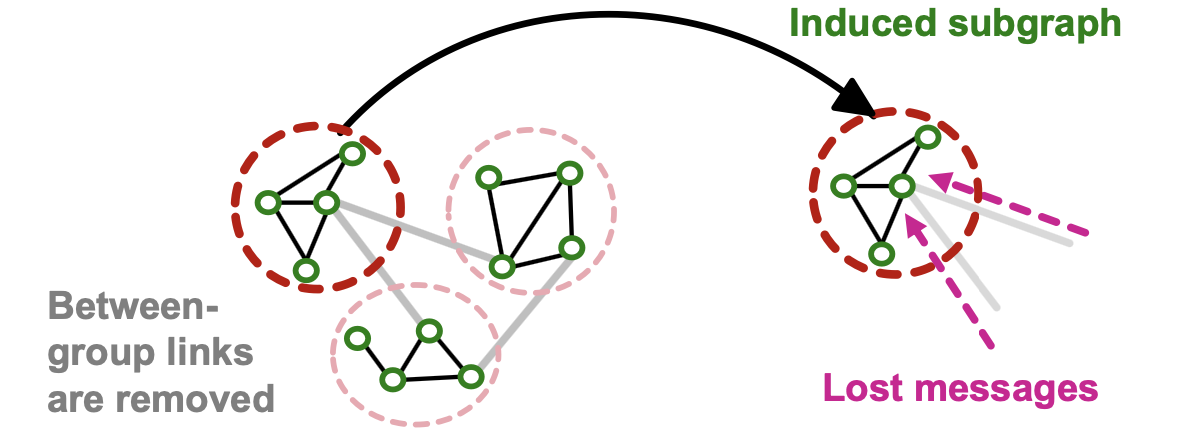
Issues with Cluster-GCN
- The induced subgraph removes between-group links.
- As a result, messages from other groups will be lot during message passing, which could hurt the GNN’s performance
- Graph community detection algorithm puts similar nodes together in the same group.
- Sampled node group tends to only cover the small-concentrated portion of the entire data.
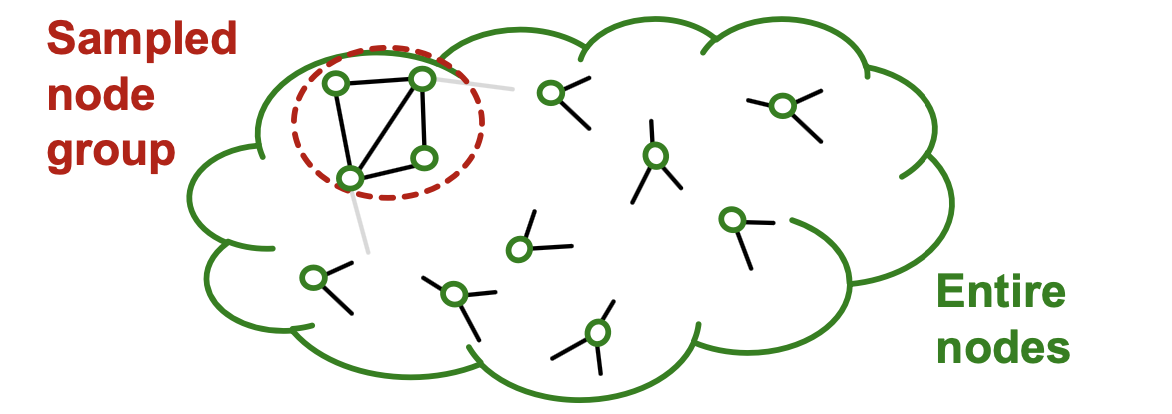
- Sampled nodes are not divers enough to be represent the entire graph structure:
- As a result, the gradient averaged over the sampled nodes, , becomes unreliable.
- Fluctuates a lot from a node group to another.
- In other words, the gradient has high variance.
- Leads to slow convergence of SGD
- As a result, the gradient averaged over the sampled nodes, , becomes unreliable.
- Solution: Aggregate multiple node groups per mini-batch.
- Partition the graph into relatively-small groups of nodes.
- For each mini-batch:
- Sample and aggregate multiple node groups.
- Construct the induced subgraph of the aggregated node group.
- The rest is the same as vanilla Cluster-GCN (compute node embeddings and the loss , update parameters)
- Why does the solution work?
Advanced Cluster-GCN
Similar to vanilla Cluster-GCN, advanced Cluster-GCN also follows a 2-step approach.
- 1) Pre-processing step:
- Given a large graph , partition its nodes into relatively-small groups:
- needs to be small so that even if multiple of them are aggregated, the resulting group would not be too large.
- Given a large graph , partition its nodes into relatively-small groups:
- 2) Mini-batch training:
- For each mini-batch, randomly sample a set of node groups: .
- Aggregate all nodes across the sampled node groups:
- Extract the induced subgraph where
- also includes between-group edges!
Comparison of Time Complexity
- Generate node embeddings using -layer GNN (N: #all nodes)
- Neighbor-sampling (sampling nodes per layer):
- For each node, the size of -layer computational graph is .
- For nodes, the cost is
- Cluster-GCN:
- Perform message passing over a subgraph induced by the nodes.
- The subgraph contains edges, where is the average node degree.
- K-layer message passing over the subgraph costs at most .
- In summary, the cost to generate embeddings for nodes using -layer GNN is:
- Neighbor-sampling (sample nodes per layer):
- Cluster-GCN:
- Assume . In other words, of neighbors are sampled.
- Then, Cluster-GCN (cost: ) is much more efficient than neighbor sampling (cost: ).
- Linear (instead of exponential) dependency w.r.t. .
Scaling up by Simplifying GNN Architecture
Roadmap of Simplifying GCN
- We start from Graph Convolutional Network (GCN)
- We simplify GCN(”SimplGCN”) by removing the non-linear activation from the GCN.
- SimplGCN demonstrated that the performance on benchmark is not much lower by the simplification.
- Simplified GCN turns out to be extremely scalable by the model design.
- The simplification strategy is very similar to the one used by LightGCN for recommender systems.
Quick Overview of LightGCN
- Adjacency matrix:
- Degree matrix:
- Normalized adjacency matrix:
- Let be the embedding matrix at -th layer.
- Let be the input embedding matrix.
- We backprop into .
- GCN’s aggregation in the matrix form
-
- Removing ReLU non-linearity gives us
- , where
- : Diffusing node embeddings along the graph.
- , where
- Efficient algorithm to obtain
- Start from input embedding matrix .
- Apply for times.
- Weight matrix can be ignored for now.
- acts as a linear classifier over the diffused node embeddings .
Differences to LightGCN
- SimplGCN adds self-loops to adjacency matrix :
-
- Follows the original GCN by Kipf & Welling.
-
- SimplGCN assumes input node embeddings to be given as features:
- Input embedding matrix is fixed rather than learned.
- Important consequence: needs to be calculated only once.
- Can be treated as a pre-processing step.
Simplified GCN: “SimplGCN”
- Let be pre-processed feature matrix.
- Each row stores the pre-processed feature for each node.
- can be used as input to any scalable ML models (e.g., linear model, MLP).
- SimplGCN empirically shows learning a linear model over often gives performance comparable to GCN!
Comparison with Other Methods
- Compared to neighbor sampling and cluster-GCN, SimplGCN is much more eddicient.
- SimplGCN computes only once at the beginning.
- The pre-processing (sparse matrix vector product,() can be performed efficiently on CPU.
- Once is obtained, getting an embedding for node only takes caonstant time!
- Just look up a row for node in .
- No need to build a computational graph or sample a subgraph.
- SimplGCN computes only once at the beginning.
- But the model is less expressive.
Potential Issue of Simplified GCN
- Compared to the original GNN models, SimplGCN’s expressive power is limited due to the lack of non-linearity in generating node embeddings.
- Surprisingly, in semi-supervised node classification benchmark, SimplGCN works comparably to the original GNNs despite being less expressive.
Graph Homophily
- Many node classification tasks exhibit homophily structure, i.e., nodes connected by edges tend to share the same target labels.
When does Simplified GCN Work?
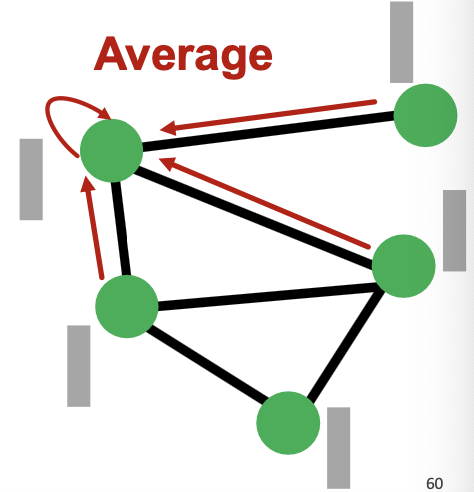
- Recall the preprocessing step of the simplified GCN: Do for times.
- is node feature matrix
- Pre-processed features are obtained by iteratively averaging their neighboring node features.
- As a result, nodes connected by edges tend to have similar pre-processed features.
- Premise: Model uses the pre-processed node features to make prediction.
- Nodes connected by edges tend to get similar pre-processed features.
- Nodes connected by edges tend to be predicted the same labels by the model
- Simplified SGC’s prediction aligns well with the graph homophily in many node classification benchmark datesets.