CS224W-Machine Learning with Graph-GNN3
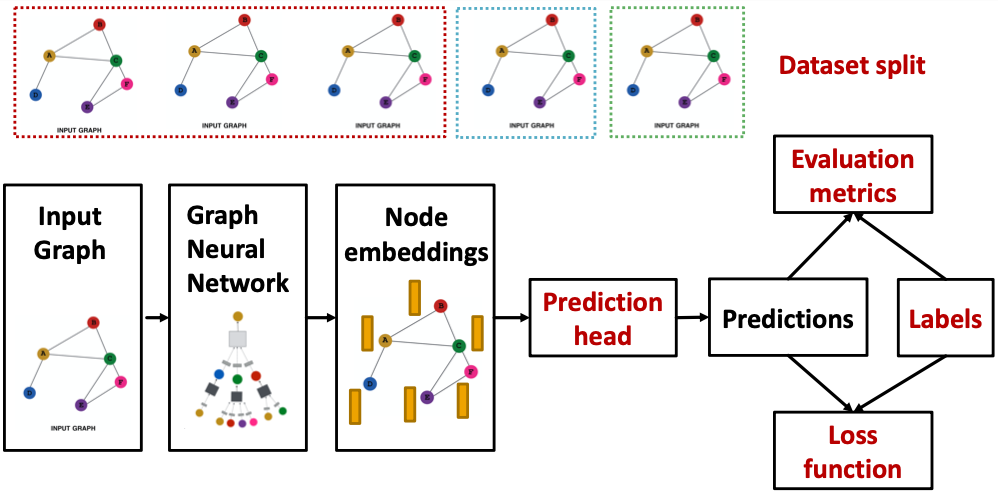
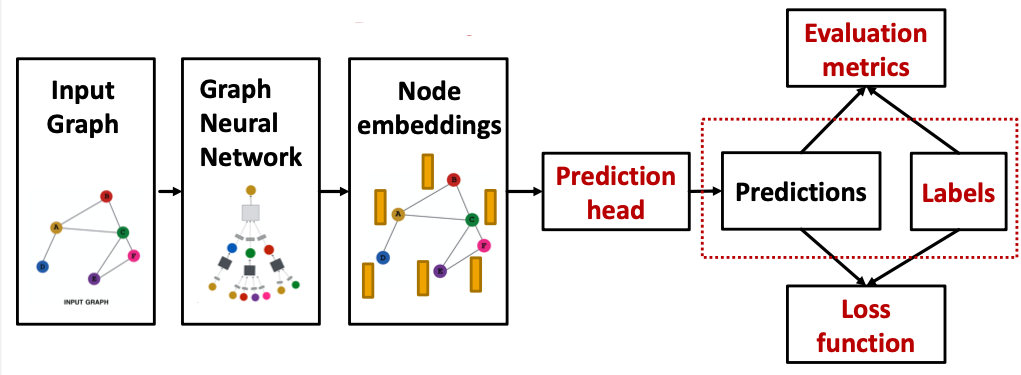
GNN Training Pipeline
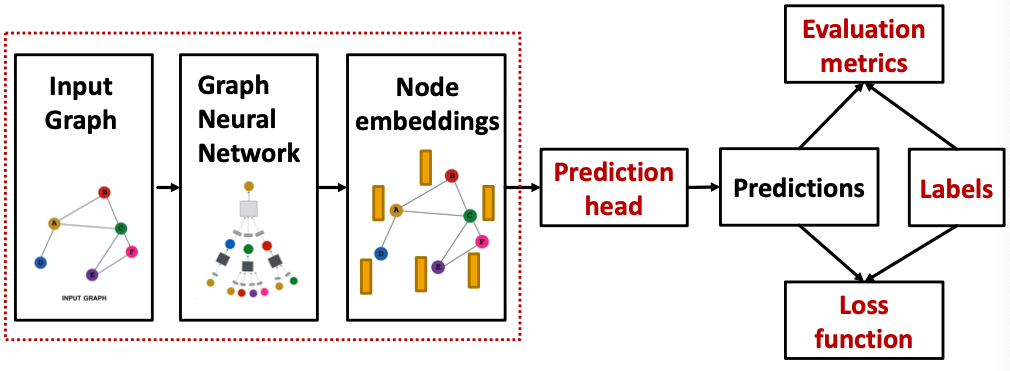
Output of a GNN: set of node embeddings
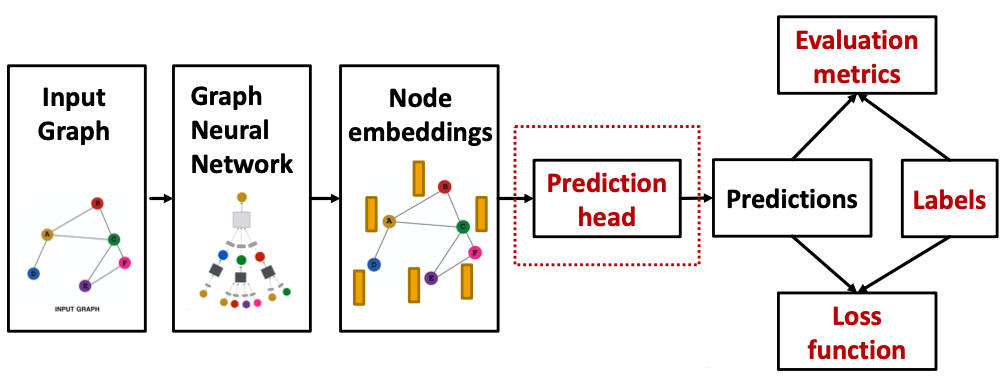
Different prediction heads: (Idea: Different task levels require different prediction heads)
- Node-level tasks
- Edge-level tasks
- Graph-level tasks
Prediction Heads: Node-level
- Node-level prediction: We can directly make prediction using node embeddings!
- After GNN computation, we have -dim node embeddings:
- Suppose we want to make -way prediction
- Classification: classify among categories
- Regression: regress on targets
-
- : We map node embeddings from to so that we can compute the loss.
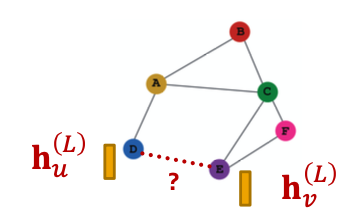
Prediction Heads: Edge-level
- Edge-level prediction: Make prediction using pairs of node embeddings
- Suppose we want to make -way prediction
- Concatenation + Linear
- We have seen this is graph attention
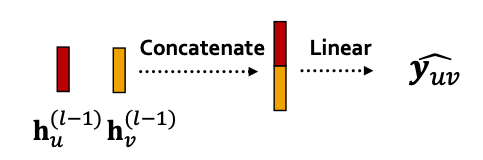
-
- Here will map -dimensional embeddings (since we concatenated embeddings) to -dim embeddings (-way prediction)
- Dot product
-
- This approach only applies to -way prediction
- Applying to -way prediction:
- Similar to multi-head attention: trainable
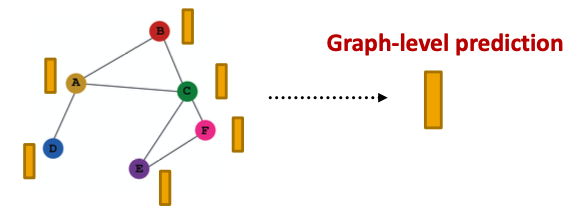
Prediction Heads: Graph-level
- Graph-level prediction: Make prediction using all the node embeddings in our graph
- Suppose we want to make -way prediction
- is similar to in a GNN layer!
- Global mean pooling
- Global max pooling
- Global sum pooling
Where does ground-truth come form?
- Supervised Labels
- Unsupervised Signals
Supervised learning on graphs
- Labels come form external sources (predict drug likeness of a molecular graph)
Unsupervised learning on graphs
- Signals come from graphs themselves (link prediction: predict if two nodes are connected)
Sometimes the differences are blurry
- We still have “supervision” in unsupervised learning (train a GNN to predict node clustering coefficient)
- An alternative name for “unsupervised” is “self-supervised”(自监督模型)
Supervised labels come from the specific use cases.
- Node labels in a citation network, which subject area does a node belong to
- Edge labels in a transaction network, whether an edge is fraudulent
- Graph labels among molecular graphs, the drug likeness of graphs
Advice: Reduce your task to node /edge/ graph labels, since they are easy to work with.
The problem: sometimes we only have a graph without any external labels
The solutions: ”self-supervised learning”, we can find supervision signals within the graph.
For example:
- Node-level Node statistics: such as clustering coefficient, PageRank,…
- Edge-level Link prediction: hidden the edge between two nodes, predict if there should be a link
- Graph-level Graph statistics: for example, predict if two graphs are isomorphic
Advice:These tasks do not require any external labels!
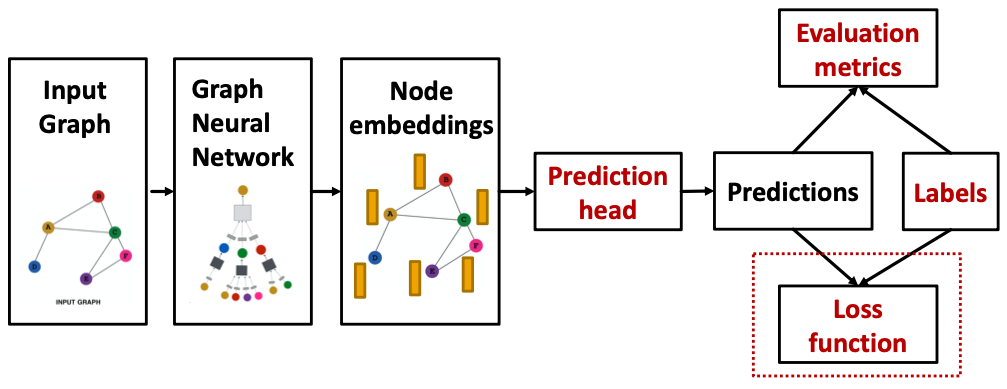
How do we compute the final loss?
- Classification loss
- Regression loss
Settings for GNN Training
We have data points
- Each data point can be a node/edge/graph
- Node-level: prediction , label
- Edge-level: prediction , label
- Graph-level: prediction , label
Classification: labels with discrete value
Regression: labels with continuous value
Classification Loss
- cross entropy (CE) is a very common loss function in classification
- -way prediction for -th data point:
where:
- Total loss over all training examples
Regression Loss
- For regression tasks we often use Mean Squared Error (MSE) a.k.a loss
- -way regression for data point :
where
- Total loss over all training examplles
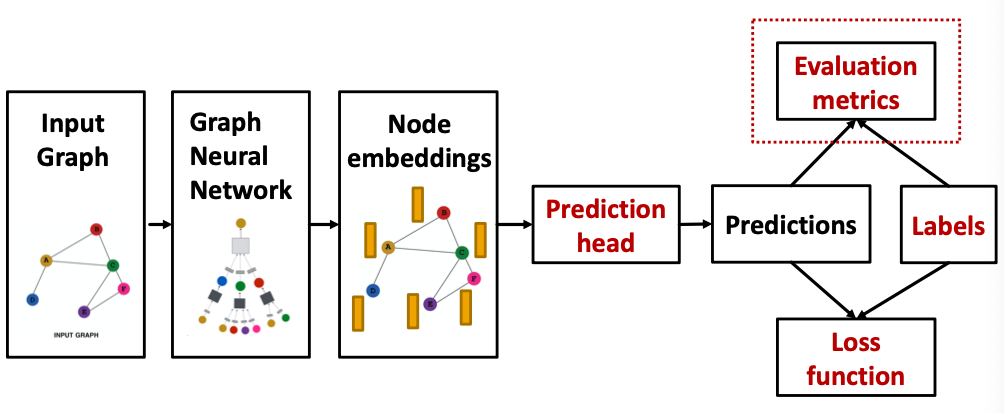
How do we measure the success of a GNN?
- Accuracy
- ROC AUC
Evaluation Metrics: Regression
Suppose we make predictions for data points
- Root mean square error (RMSE)
- Mean absolute error (MAE)
Evaluation Metrics: Classification
- Multi-class classification
We simply report the accuracy
- Binary classification
- Metrics sensitive to classification threshold
- Accuracy
- Precision / Recall
- If the range of prediction is , we will use as threshold
- Metric Agnostic to classification threshold
- ROC AUC
- Metrics sensitive to classification threshold
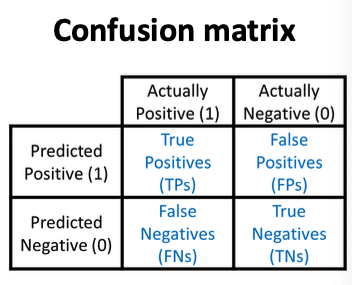
Accuracy
Precision (P)
Recall (R)
Score
ROC Curve: Captures the tradeoff in TPR and FPR as the classification threshold is varied for a binary classifier.
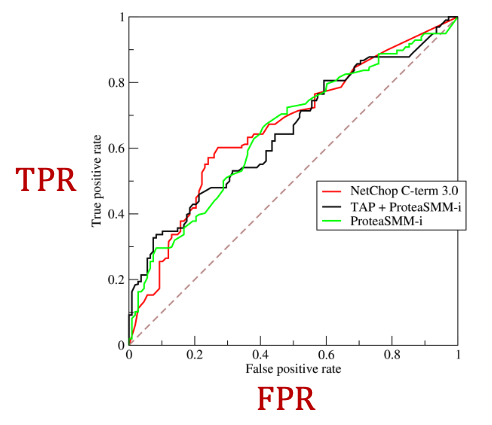
ROC AUC: Area under the ROC Curve.
Intuition: The probability that a classifier will rank a randomly chosen positive instance higher than a randomly chosen negative one.
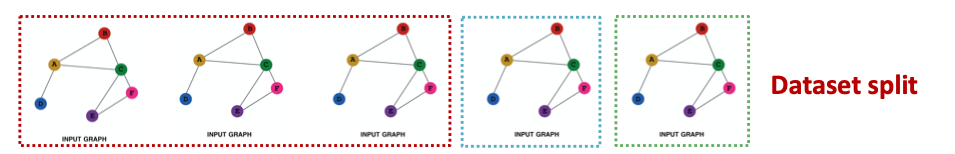
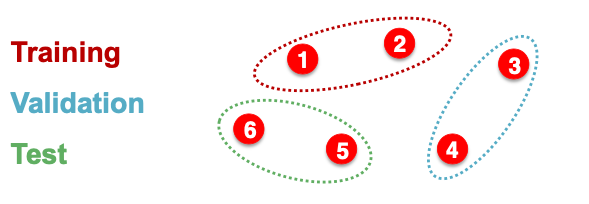
How do we split our dataset into train / validation / test set?
Dataset Split: Fixed / Random Split
- Fixed split: we will split our dataset once
- Training set: used for optimizing GNN parameters
- Validation set: develop model / hyperparameters
- Test set: held out until we report final performance
- A concern: sometimes we cannot guarantee that the test set will really be held out
- Random split: we will randomly split out dataset into training / validation / test
- We report average performance over different random seed.
分割图是特殊的 为什么? Why Splitting Graphs is Special? 或许每个节点都有可能是相互关联的
Suppose we want to split an image dataset?
- Image classification: Each data point is an image
Splitting a graph dataset is different!
- Node classification: Each data point is a node
- Here data points are NOT independent
- Node 5 will affect our prediction on node 1, because it will participate in message passing affect node 1’s embedding
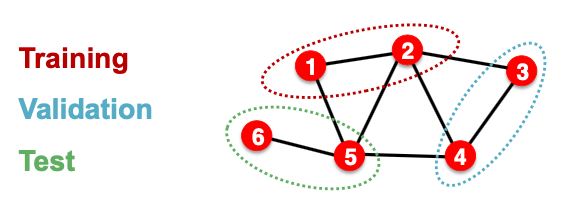
The input graph can be observed in all the dataset splits (training, validation and test set)
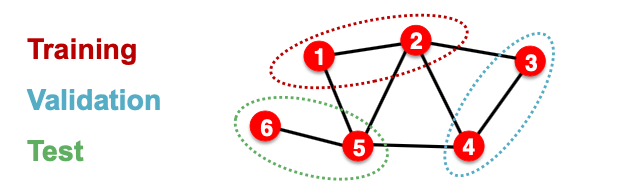
- We will only split the (node) labels
- At training time, we compute embeddings using the entire graph, and train using node 1&2’s labels
- At validation time, we compute embeddings using the entire graph, and evaluate on node 3&4’s labels
We break the edges between splits to get multiple graphs
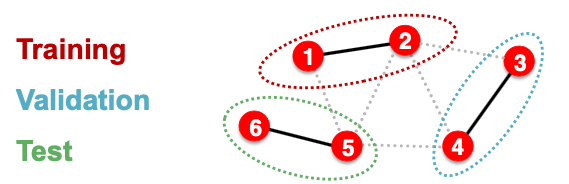
- Now we have 3 graphs that are independent. Node 5 will not affect our prediction on node 1 any more.
- At training time, we compute embeddings using the graph over node 1&2, and train using node 1&2’s labels
- At validation time, we compute embeddings using the graph over node 3&4, and evaluate on node 3&4’s labels
Transductive / Inductive Settings
- Transductive setting: training / validation / test sets are on the same graph
- The dataset consists of one graph
- The entire graph can be observed in all dataset splits, we only split the labels
- Only applicable to node /edge prediction tasks
- Inductive setting: training / validation / test sets are on different graphs
- The dataset consists of multiple graphs
- Each split can only observe the graph within the split. A successful model should generalize to unseen graphs
- Applicable to node /edge / graph tasks
- Transductive node classification
- All the splits can observe the entire graph structure, but can only observe the labels of their respective nodes
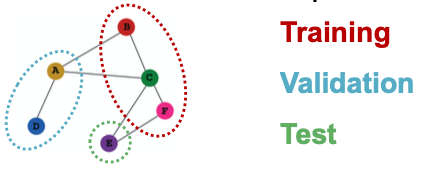
- Inductive node classification
- Suppose we have a dataset of 3 graphs
- Each split contains an independent graph
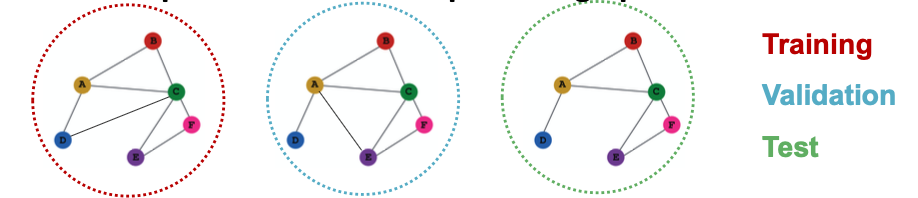
- Only the inductive setting is well defined for graph classification
- Because we have to test on unseen graphs
- Suppose we have a dataset of 5 graphs. Each split will contain independent graph(s).
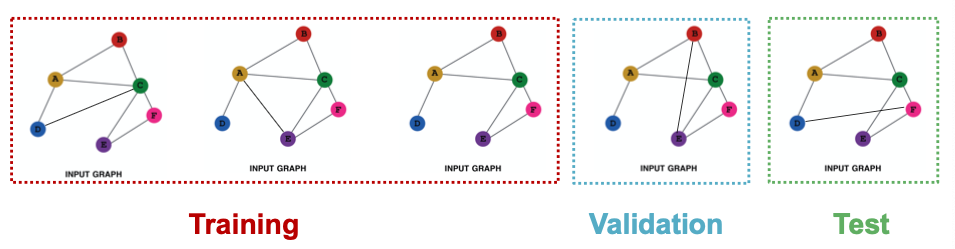
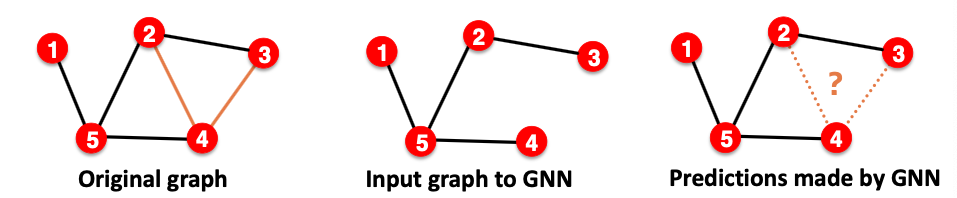
- Goal of link prediction: predict missing edges
- Setting up link prediction is tricky(确实):
- Link prediction is an unsupervised / self-supervised task. We need to create the labels and dataset splits on our own.
- Concretely, we need to hide some edges from the GNN and the let the GNN predict if the edges exist.
How to set up link prediction?
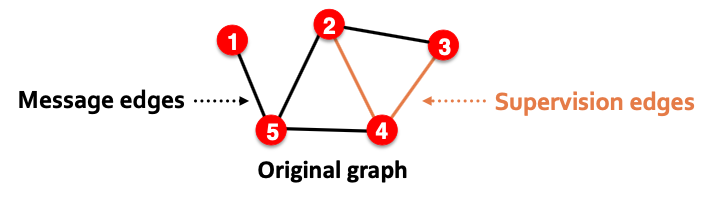
- For link prediction, we will split edges twice
- Step 1: Assign 2 types of edges in the original graph
- Message edges: Used for GNN message passing
- Supervision edges: Use for computing objectives
- After step 1:
- Only message edges will remain in the graph
- Supervision edges are used as supervision for edge predictions made by the model, will not be fed into GNN!
- Step 2: Split edges into train / validation / test
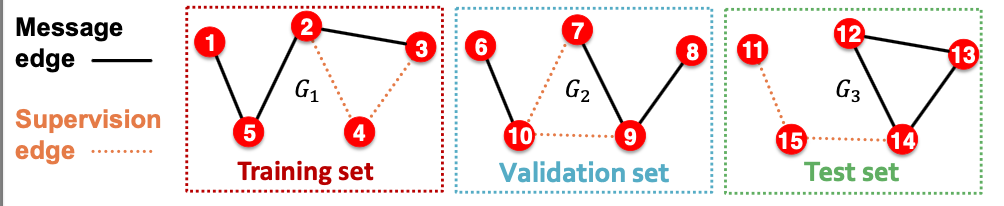
- Option 1: Inductive link prediction split
- Suppose we have a dataset of 3 graphs. Each inductive split will contain an independent graph.
- In train or val or test set, each graph will have 2 types of edges: message edges + supervision edges
- Supervision edges are not the input to GNN
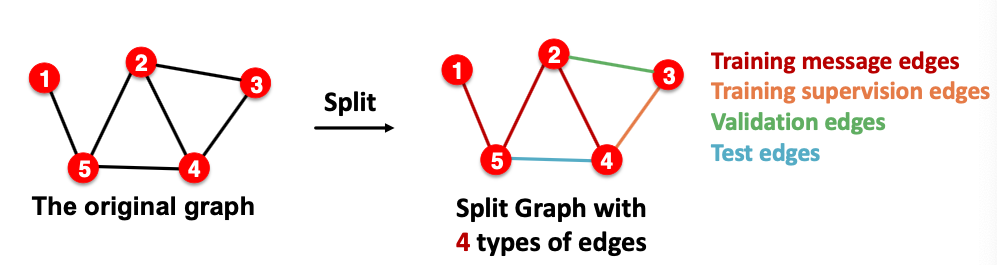
- Option 2: Transductive link prediction split
- This is default setting when people talk about link prediction
- Suppose we have a dataset of 1 graph
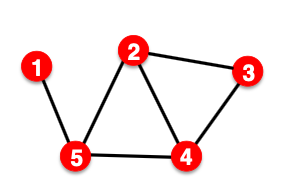
- By definition of “transductive”, the entire graph can be observed in all dataset splits
- But since edges are both part of graph structure and the supervision, we need to hold out validation / test edges
- To train the training set, we further need to hold out supervision edges for the training set
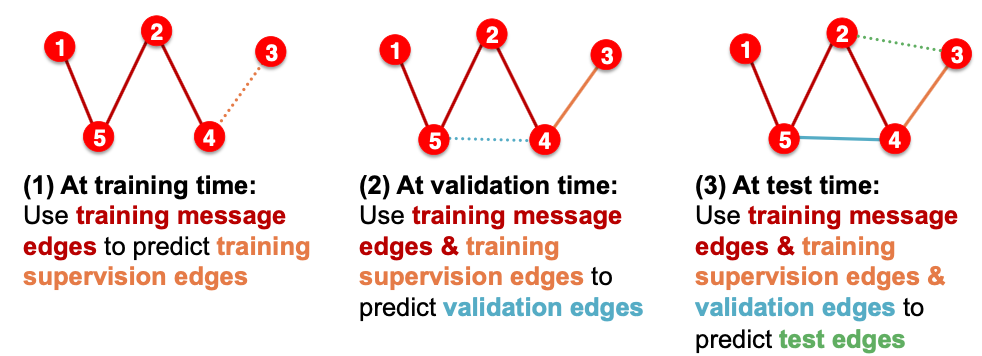
Why do we use growing number of edges?
After training, supervision edges are know to GNN. Therefore, an ideal model should use supervision edges in message passing at validation time. The same applies to the test time.
Summary: Transductive link prediction split